Christopher James Langmead
Structure-Aligned Protein Language Model
May 22, 2025Abstract:Protein language models (pLMs) pre-trained on vast protein sequence databases excel at various downstream tasks but lack the structural knowledge essential for many biological applications. To address this, we integrate structural insights from pre-trained protein graph neural networks (pGNNs) into pLMs through a latent-level contrastive learning task. This task aligns residue representations from pLMs with those from pGNNs across multiple proteins, enriching pLMs with inter-protein structural knowledge. Additionally, we incorporate a physical-level task that infuses intra-protein structural knowledge by optimizing pLMs to predict structural tokens. The proposed dual-task framework effectively incorporates both inter-protein and intra-protein structural knowledge into pLMs. Given the variability in the quality of protein structures in PDB, we further introduce a residue loss selection module, which uses a small model trained on high-quality structures to select reliable yet challenging residue losses for the pLM to learn. Applying our structure alignment method to the state-of-the-art ESM2 and AMPLIFY results in notable performance gains across a wide range of tasks, including a 12.7% increase in ESM2 contact prediction. The data, code, and resulting SaESM2 and SaAMPLIFY models will be released on Hugging Face.
Peptide Binding Classification on Quantum Computers
Nov 27, 2023



Abstract:We conduct an extensive study on using near-term quantum computers for a task in the domain of computational biology. By constructing quantum models based on parameterised quantum circuits we perform sequence classification on a task relevant to the design of therapeutic proteins, and find competitive performance with classical baselines of similar scale. To study the effect of noise, we run some of the best-performing quantum models with favourable resource requirements on emulators of state-of-the-art noisy quantum processors. We then apply error mitigation methods to improve the signal. We further execute these quantum models on the Quantinuum H1-1 trapped-ion quantum processor and observe very close agreement with noiseless exact simulation. Finally, we perform feature attribution methods and find that the quantum models indeed identify sensible relationships, at least as well as the classical baselines. This work constitutes the first proof-of-concept application of near-term quantum computing to a task critical to the design of therapeutic proteins, opening the route toward larger-scale applications in this and related fields, in line with the hardware development roadmaps of near-term quantum technologies.
Deep Normed Embeddings for Patient Representation
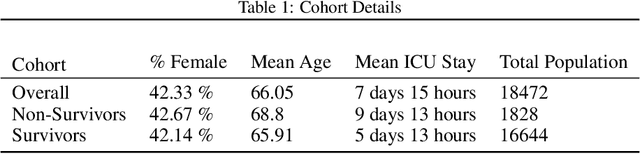
Apr 12, 2022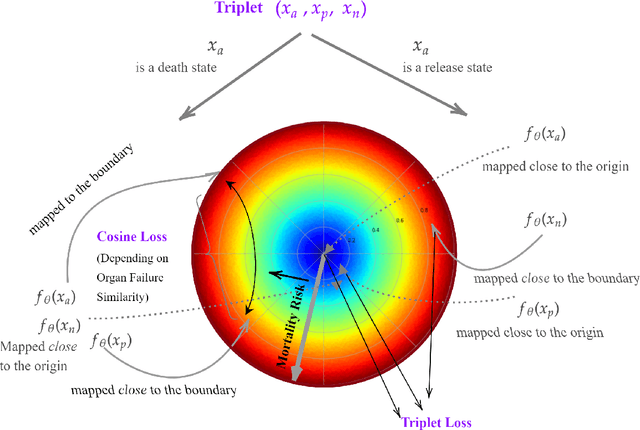
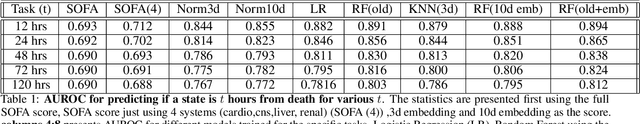
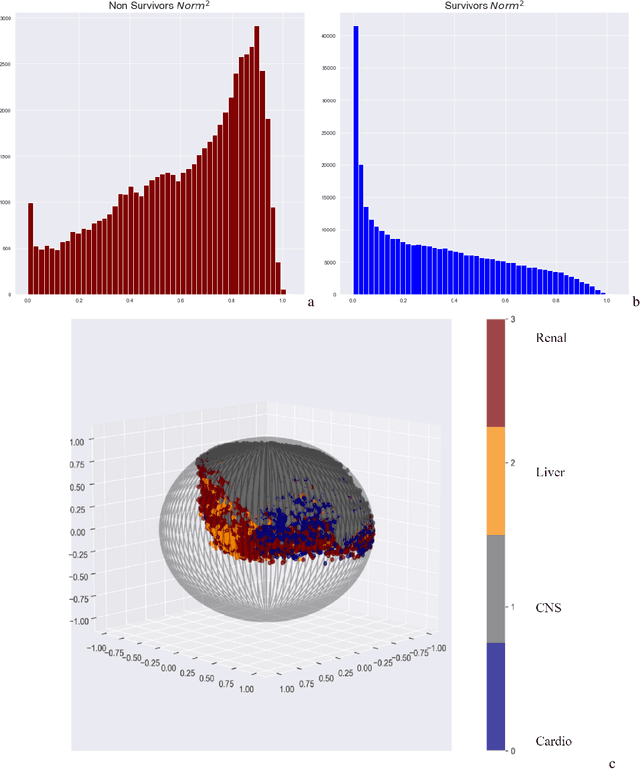

Abstract:We introduce a novel contrastive representation learning objective and a training scheme for clinical time series. Specifically, we project high dimensional E.H.R. data to a closed unit ball of low dimension, encoding geometric priors so that the origin represents an idealized perfect health state and the euclidean norm is associated with the patient's mortality risk. Moreover, using septic patients as an example, we show how we could learn to associate the angle between two vectors with the different organ system failures, thereby, learning a compact representation which is indicative of both mortality risk and specific organ failure. We show how the learned embedding can be used for online patient monitoring, supplement clinicians and improve performance of downstream machine learning tasks. This work was partially motivated from the desire and the need to introduce a systematic way of defining intermediate rewards for Reinforcement Learning in critical care medicine. Hence, we also show how such a design in terms of the learned embedding can result in qualitatively different policies and value distributions, as compared with using only terminal rewards.
Unifying Cardiovascular Modelling with Deep Reinforcement Learning for Uncertainty Aware Control of Sepsis Treatment
Feb 02, 2021


Abstract:Sepsis is the leading cause of mortality in the ICU, responsible for 6% of all hospitalizations and 35% of all in-hospital deaths in USA. However, there is no universally agreed upon strategy for vasopressor and fluid administration. It has also been observed that different patients respond differently to treatment, highlighting the need for individualized treatment. Vasopressors and fluids are administrated with specific effects to cardiovascular physiology in mind and medical research has suggested that physiologic, hemodynamically guided, approaches to treatment. Thus we propose a novel approach, exploiting and unifying complementary strengths of Mathematical Modelling, Deep Learning, Reinforcement Learning and Uncertainty Quantification, to learn individualized, safe, and uncertainty aware treatment strategies. We first infer patient-specific, dynamic cardiovascular states using a novel physiology-driven recurrent neural network trained in an unsupervised manner. This information, along with a learned low dimensional representation of the patient's lab history and observable data, is then used to derive value distributions using Batch Distributional Reinforcement Learning. Moreover in a safety critical domain it is essential to know what our agent does and does not know, for this we also quantify the model uncertainty associated with each patient state and action, and propose a general framework for uncertainty aware, interpretable treatment policies. This framework can be tweaked easily, to reflect a clinician's own confidence of the framework, and can be easily modified to factor in human expert opinion, whenever it's accessible. Using representative patients and a validation cohort, we show that our method has learned physiologically interpretable generalizable policies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge