Christoph Baur
Scale-Space Autoencoders for Unsupervised Anomaly Segmentation in Brain MRI
Jun 23, 2020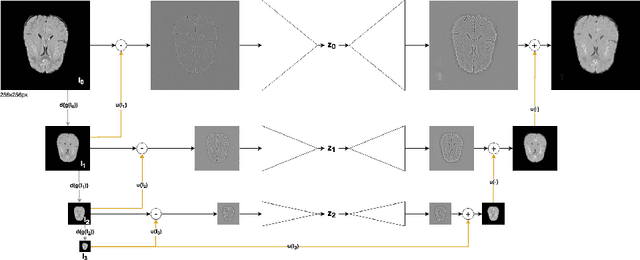


Abstract:Brain pathologies can vary greatly in size and shape, ranging from few pixels (i.e. MS lesions) to large, space-occupying tumors. Recently proposed Autoencoder-based methods for unsupervised anomaly segmentation in brain MRI have shown promising performance, but face difficulties in modeling distributions with high fidelity, which is crucial for accurate delineation of particularly small lesions. Here, similar to these previous works, we model the distribution of healthy brain MRI to localize pathologies from erroneous reconstructions. However, to achieve improved reconstruction fidelity at higher resolutions, we learn to compress and reconstruct different frequency bands of healthy brain MRI using the laplacian pyramid. In a range of experiments comparing our method to different State-of-the-Art approaches on three different brain MR datasets with MS lesions and tumors, we show improved anomaly segmentation performance and the general capability to obtain much more crisp reconstructions of input data at native resolution. The modeling of the laplacian pyramid further enables the delineation and aggregation of lesions at multiple scales, which allows to effectively cope with different pathologies and lesion sizes using a single model.
Autoencoders for Unsupervised Anomaly Segmentation in Brain MR Images: A Comparative Study
Apr 08, 2020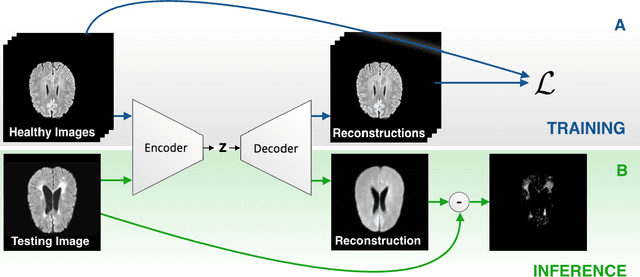
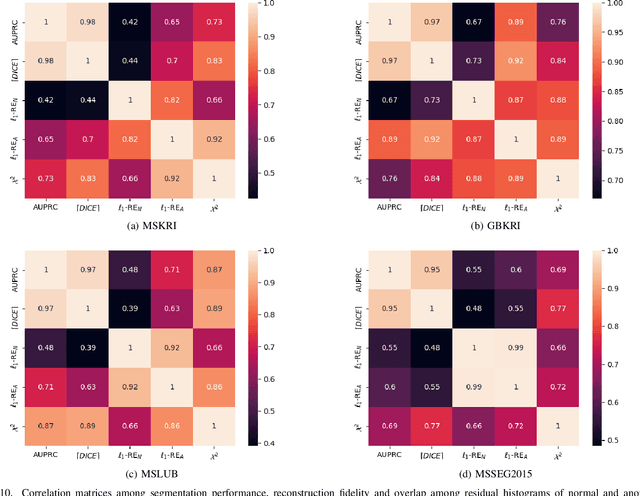
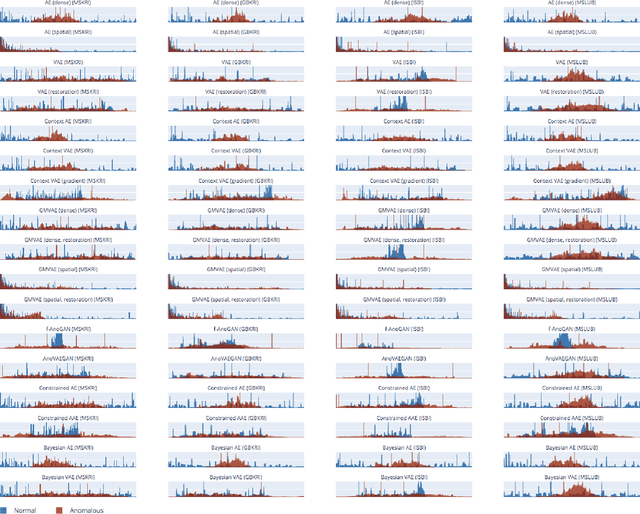
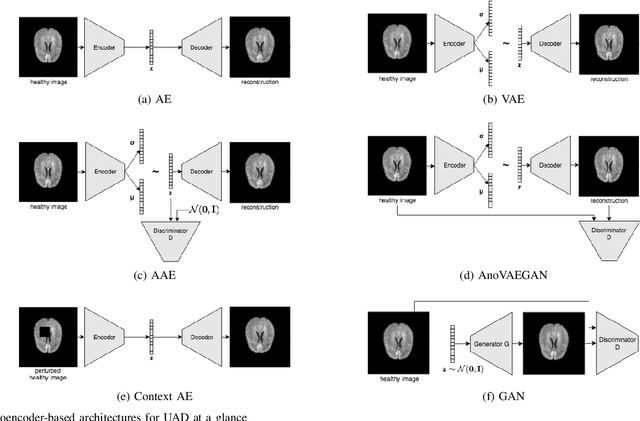
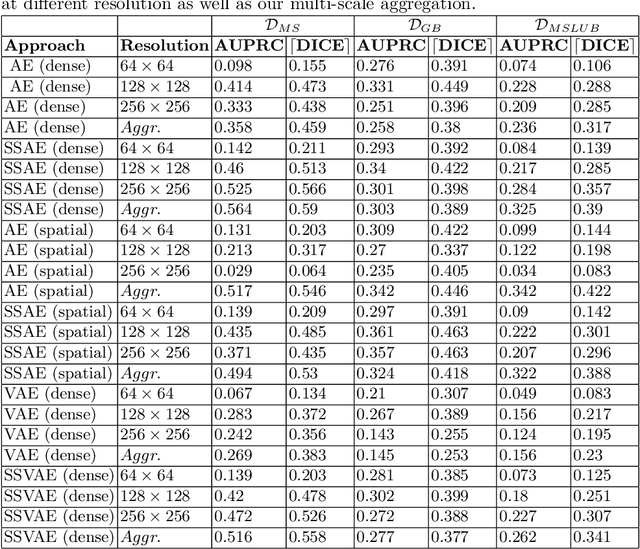
Abstract:Deep unsupervised representation learning has recently led to new approaches in the field of Unsupervised Anomaly Detection (UAD) in brain MRI. The main principle behind these works is to learn a model of normal anatomy by learning to compress and recover healthy data. This allows to spot abnormal structures from erroneous recoveries of compressed, potentially anomalous samples. The concept is of great interest to the medical image analysis community as it i) relieves from the need of vast amounts of manually segmented training data---a necessity for and pitfall of current supervised Deep Learning---and ii) theoretically allows to detect arbitrary, even rare pathologies which supervised approaches might fail to find. To date, the experimental design of most works hinders a valid comparison, because i) they are evaluated against different datasets and different pathologies, ii) use different image resolutions and iii) different model architectures with varying complexity. The intent of this work is to establish comparability among recent methods by utilizing a single architecture, a single resolution and the same dataset(s). Besides providing a ranking of the methods, we also try to answer questions like i) how many healthy training subjects are needed to model normality and ii) if the reviewed approaches are also sensitive to domain shift. Further, we identify open challenges and provide suggestions for future community efforts and research directions.
Adversarial Joint Image and Pose Distribution Learning for Camera Pose Regression and Refinement
Mar 26, 2019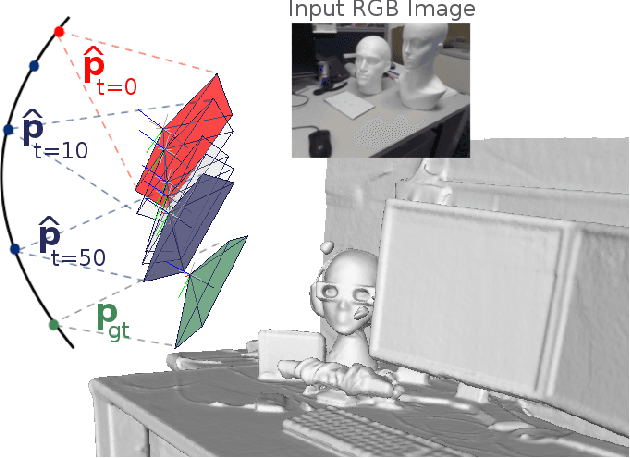
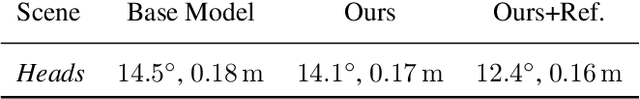
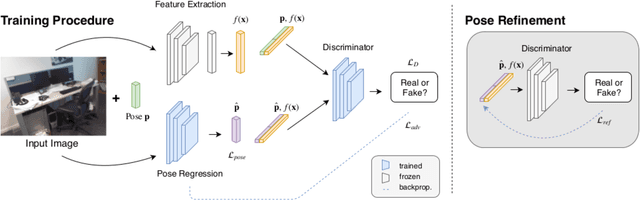

Abstract:Despite recent advances on the topic of direct camera pose regression using neural networks, accurately estimating the camera pose of a single RGB image still remains a challenging task. To address this problem, we introduce a novel framework based, in its core, on the idea of modeling the joint distribution of RGB images and their corresponding camera poses using adversarial learning. Our method allows not only to regress the camera pose from a single image, however, also offers a solely RGB-based solution for camera pose refinement using the discriminator network. Further, we show that our method can effectively be used to optimize the predicted camera poses and thus improve the localization accuracy. To this end, we validate our proposed method on the publicly available 7-Scenes dataset improving upon the results of current state-of-the-art direct camera pose regression methods.
GANs for Medical Image Analysis
Sep 13, 2018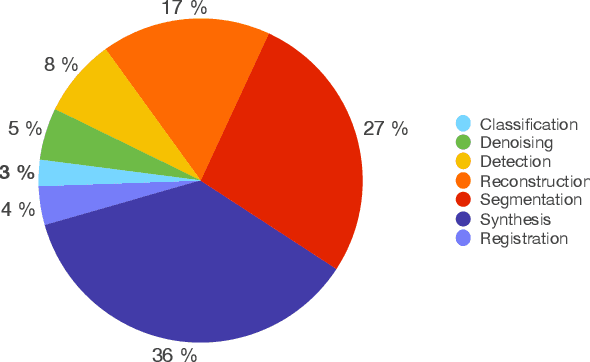
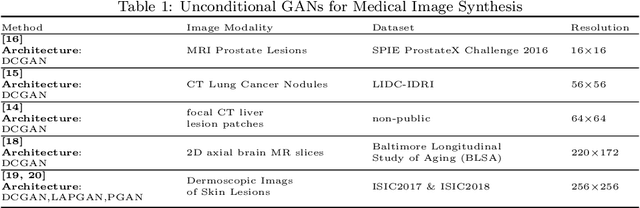
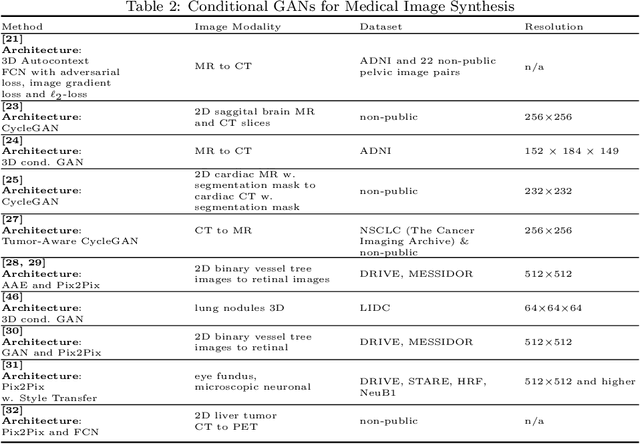
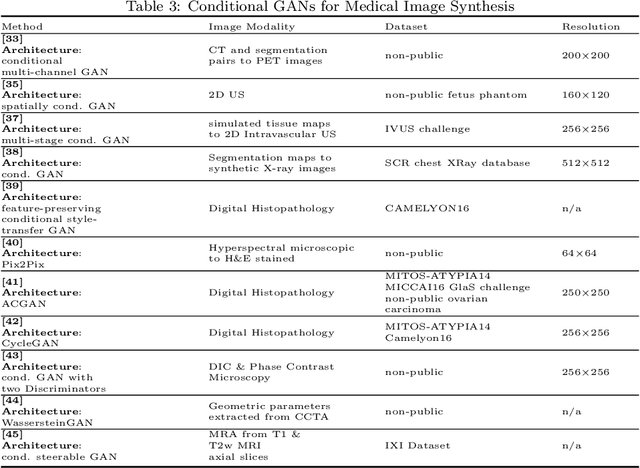
Abstract:Generative Adversarial Networks (GANs) and their extensions have carved open many exciting ways to tackle well known and challenging medical image analysis problems such as medical image denoising, reconstruction, segmentation, data simulation, detection or classification. Furthermore, their ability to synthesize images at unprecedented levels of realism also gives hope that the chronic scarcity of labeled data in the medical field can be resolved with the help of these generative models. In this review paper, a broad overview of recent literature on GANs for medical applications is given, the shortcomings and opportunities of the proposed methods are thoroughly discussed and potential future work is elaborated. A total of 63 papers published until end of July 2018 are reviewed. For quick access, the papers and important details such as the underlying method, datasets and performance are summarized in tables.
Generating Highly Realistic Images of Skin Lesions with GANs
Sep 06, 2018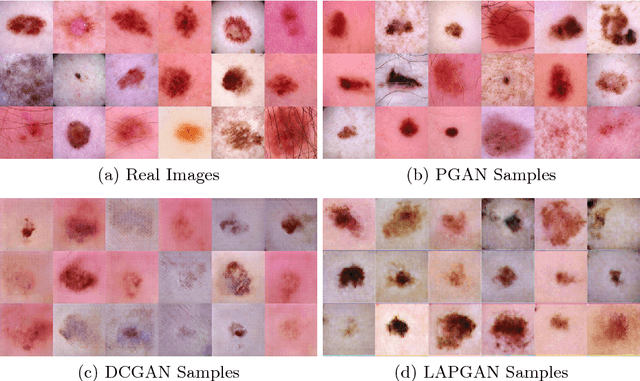


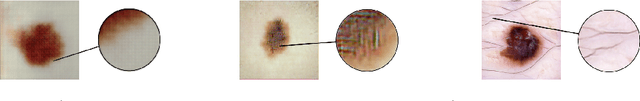
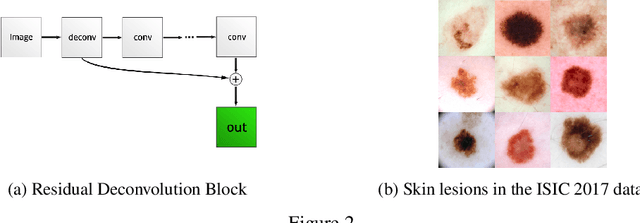
Abstract:As many other machine learning driven medical image analysis tasks, skin image analysis suffers from a chronic lack of labeled data and skewed class distributions, which poses problems for the training of robust and well-generalizing models. The ability to synthesize realistic looking images of skin lesions could act as a reliever for the aforementioned problems. Generative Adversarial Networks (GANs) have been successfully used to synthesize realistically looking medical images, however limited to low resolution, whereas machine learning models for challenging tasks such as skin lesion segmentation or classification benefit from much higher resolution data. In this work, we successfully synthesize realistically looking images of skin lesions with GANs at such high resolution. Therefore, we utilize the concept of progressive growing, which we both quantitatively and qualitatively compare to other GAN architectures such as the DCGAN and the LAPGAN. Our results show that with the help of progressive growing, we can synthesize highly realistic dermoscopic images of skin lesions that even expert dermatologists find hard to distinguish from real ones.
Deep Autoencoding Models for Unsupervised Anomaly Segmentation in Brain MR Images
Apr 12, 2018



Abstract:Reliably modeling normality and differentiating abnormal appearances from normal cases is a very appealing approach for detecting pathologies in medical images. A plethora of such unsupervised anomaly detection approaches has been made in the medical domain, based on statistical methods, content-based retrieval, clustering and recently also deep learning. Previous approaches towards deep unsupervised anomaly detection model patches of normal anatomy with variants of Autoencoders or GANs, and detect anomalies either as outliers in the learned feature space or from large reconstruction errors. In contrast to these patch-based approaches, we show that deep spatial autoencoding models can be efficiently used to capture normal anatomical variability of entire 2D brain MR images. A variety of experiments on real MR data containing MS lesions corroborates our hypothesis that we can detect and even delineate anomalies in brain MR images by simply comparing input images to their reconstruction. Results show that constraints on the latent space and adversarial training can further improve the segmentation performance over standard deep representation learning.
MelanoGANs: High Resolution Skin Lesion Synthesis with GANs
Apr 12, 2018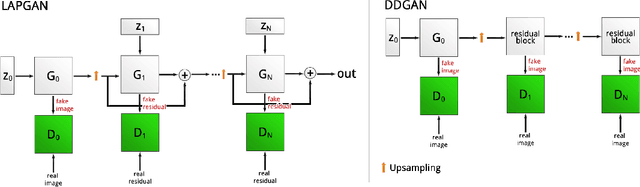
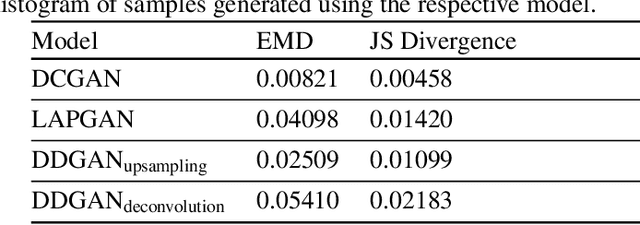

Abstract:Generative Adversarial Networks (GANs) have been successfully used to synthesize realistically looking images of faces, scenery and even medical images. Unfortunately, they usually require large training datasets, which are often scarce in the medical field, and to the best of our knowledge GANs have been only applied for medical image synthesis at fairly low resolution. However, many state-of-the-art machine learning models operate on high resolution data as such data carries indispensable, valuable information. In this work, we try to generate realistically looking high resolution images of skin lesions with GANs, using only a small training dataset of 2000 samples. The nature of the data allows us to do a direct comparison between the image statistics of the generated samples and the real dataset. We both quantitatively and qualitatively compare state-of-the-art GAN architectures such as DCGAN and LAPGAN against a modification of the latter for the task of image generation at a resolution of 256x256px. Our investigation shows that we can approximate the real data distribution with all of the models, but we notice major differences when visually rating sample realism, diversity and artifacts. In a set of use-case experiments on skin lesion classification, we further show that we can successfully tackle the problem of heavy class imbalance with the help of synthesized high resolution melanoma samples.
StainGAN: Stain Style Transfer for Digital Histological Images
Apr 04, 2018

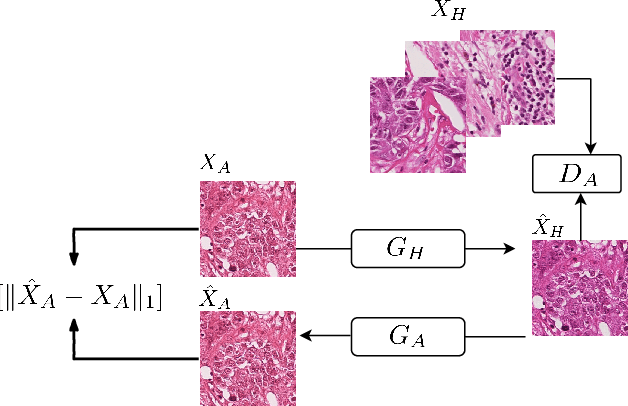
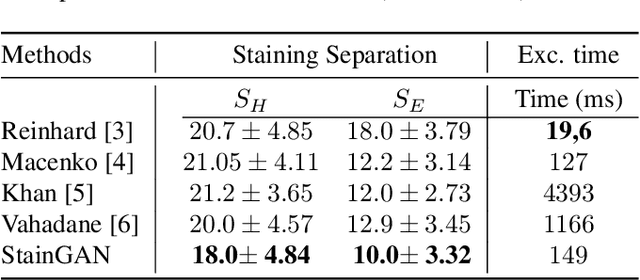
Abstract:Digitized Histological diagnosis is in increasing demand. However, color variations due to various factors are imposing obstacles to the diagnosis process. The problem of stain color variations is a well-defined problem with many proposed solutions. Most of these solutions are highly dependent on a reference template slide. We propose a deep-learning solution inspired by CycleGANs that is trained end-to-end, eliminating the need for an expert to pick a representative reference slide. Our approach showed superior results quantitatively and qualitatively against the state of the art methods (10% improvement visually using SSIM). We further validated our method on a clinical use-case, namely Breast Cancer tumor classification, showing 12% increase in AUC. The code will be made publicly available.
Semi-Supervised Deep Learning for Fully Convolutional Networks
Jul 25, 2017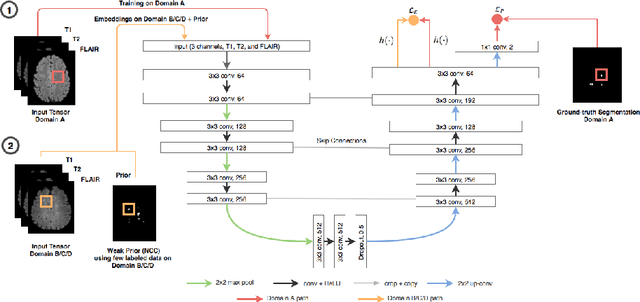

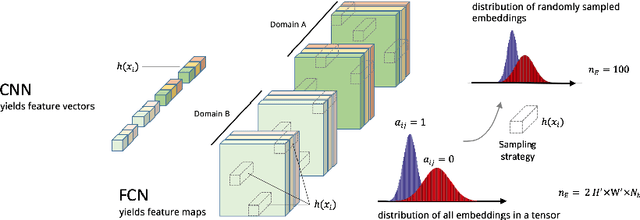

Abstract:Deep learning usually requires large amounts of labeled training data, but annotating data is costly and tedious. The framework of semi-supervised learning provides the means to use both labeled data and arbitrary amounts of unlabeled data for training. Recently, semi-supervised deep learning has been intensively studied for standard CNN architectures. However, Fully Convolutional Networks (FCNs) set the state-of-the-art for many image segmentation tasks. To the best of our knowledge, there is no existing semi-supervised learning method for such FCNs yet. We lift the concept of auxiliary manifold embedding for semi-supervised learning to FCNs with the help of Random Feature Embedding. In our experiments on the challenging task of MS Lesion Segmentation, we leverage the proposed framework for the purpose of domain adaptation and report substantial improvements over the baseline model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge