Cassio P. de Campos
Joints in Random Forests
Jul 11, 2020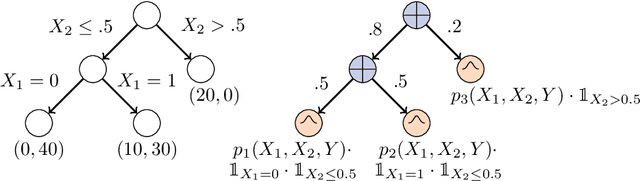
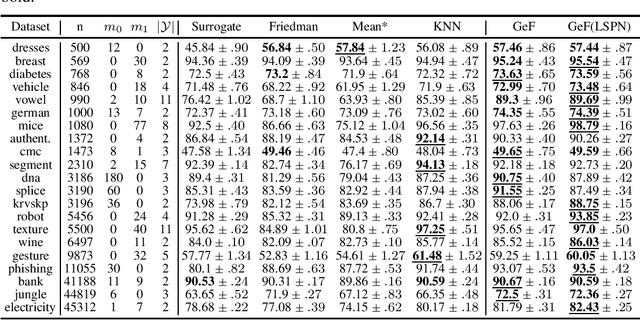
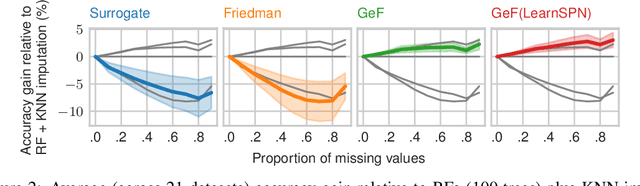
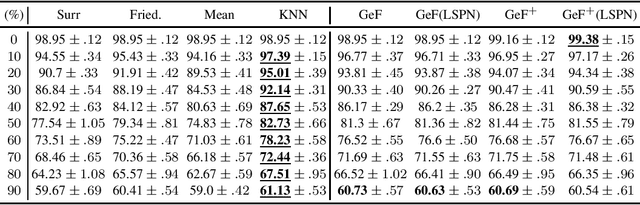
Abstract:Decision Trees (DTs) and Random Forests (RFs) are powerful discriminative learners and tools of central importance to the everyday machine learning practitioner and data scientist. Due to their discriminative nature, however, they lack principled methods to process inputs with missing features or to detect outliers, which requires pairing them with imputation techniques or a separate generative model. In this paper, we demonstrate that DTs and RFs can naturally be interpreted as generative models, by drawing a connection to Probabilistic Circuits, a prominent class of tractable probabilistic models. This reinterpretation equips them with a full joint distribution over the feature space and leads to Generative Decision Trees (GeDTs) and Generative Forests (GeFs), a family of novel hybrid generative-discriminative models. This family of models retains the overall characteristics of DTs and RFs while additionally being able to handle missing features by means of marginalisation. Under certain assumptions, frequently made for Bayes consistency results, we show that consistency in GeDTs and GeFs extend to any pattern of missing input features, if missing at random. Empirically, we show that our models often outperform common routines to treat missing data, such as K-nearest neighbour imputation, and moreover, that our models can naturally detect outliers by monitoring the marginal probability of input features.
On Pruning for Score-Based Bayesian Network Structure Learning
May 23, 2019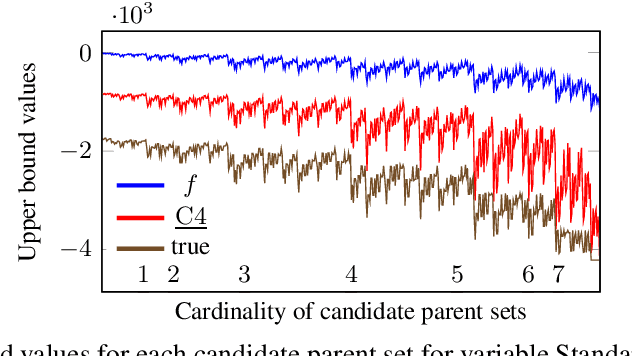
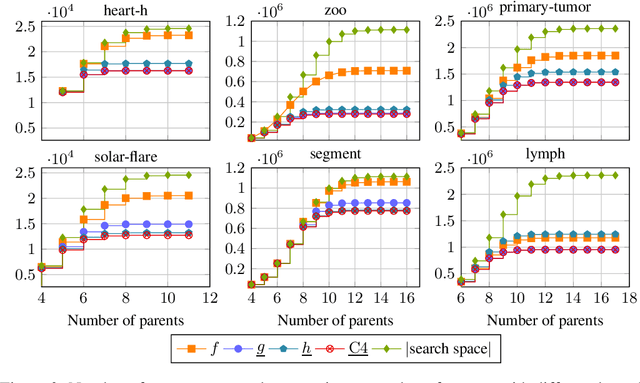
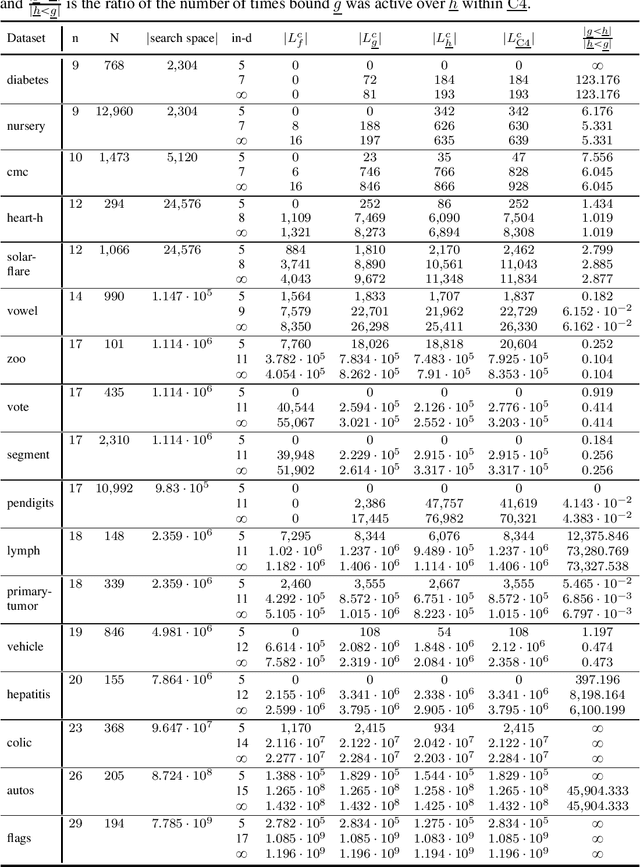
Abstract:Many algorithms for score-based Bayesian network structure learning (BNSL) take as input a collection of potentially optimal parent sets for each variable in a data set. Constructing these collections naively is computationally intensive since the number of parent sets grows exponentially with the number of variables. Therefore, pruning techniques are not only desirable but essential. While effective pruning exists for the Bayesian Information Criterion (BIC), current results for the Bayesian Dirichlet equivalent uniform (BDeu) score reduce the search space very modestly, hampering the use of (the often preferred) BDeu. We derive new non-trivial theoretical upper bounds for the BDeu score that considerably improve on the state of the art. Since the new bounds are efficient and easy to implement, they can be promptly integrated into many BNSL methods. We show that gains can be significant in multiple UCI data sets so as to highlight practical implications of the theoretical advances.
Approximation Complexity of Maximum A Posteriori Inference in Sum-Product Networks
Sep 05, 2017
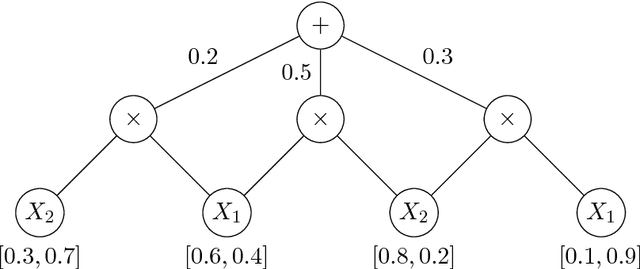

Abstract:We discuss the computational complexity of approximating maximum a posteriori inference in sum-product networks. We first show NP-hardness in trees of height two by a reduction from maximum independent set; this implies non-approximability within a sublinear factor. We show that this is a tight bound, as we can find an approximation within a linear factor in networks of height two. We then show that, in trees of height three, it is NP-hard to approximate the problem within a factor $2^{f(n)}$ for any sublinear function $f$ of the size of the input $n$. Again, this bound is tight, as we prove that the usual max-product algorithm finds (in any network) approximations within factor $2^{c \cdot n}$ for some constant $c < 1$. Last, we present a simple algorithm, and show that it provably produces solutions at least as good as, and potentially much better than, the max-product algorithm. We empirically analyze the proposed algorithm against max-product using synthetic and realistic networks.
Entropy-based Pruning for Learning Bayesian Networks using BIC
Jul 19, 2017



Abstract:For decomposable score-based structure learning of Bayesian networks, existing approaches first compute a collection of candidate parent sets for each variable and then optimize over this collection by choosing one parent set for each variable without creating directed cycles while maximizing the total score. We target the task of constructing the collection of candidate parent sets when the score of choice is the Bayesian Information Criterion (BIC). We provide new non-trivial results that can be used to prune the search space of candidate parent sets of each node. We analyze how these new results relate to previous ideas in the literature both theoretically and empirically. We show in experiments with UCI data sets that gains can be significant. Since the new pruning rules are easy to implement and have low computational costs, they can be promptly integrated into all state-of-the-art methods for structure learning of Bayesian networks.
Learning Bayesian Networks with Incomplete Data by Augmentation
Oct 09, 2016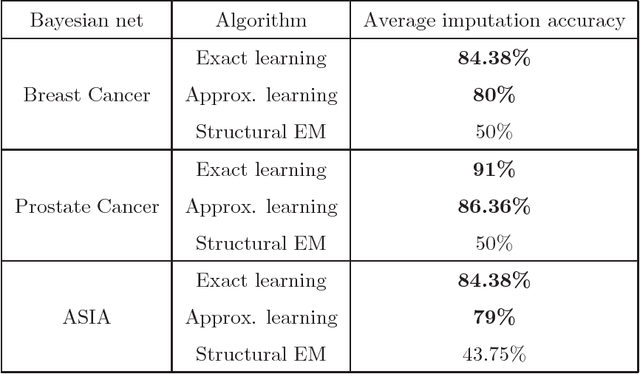
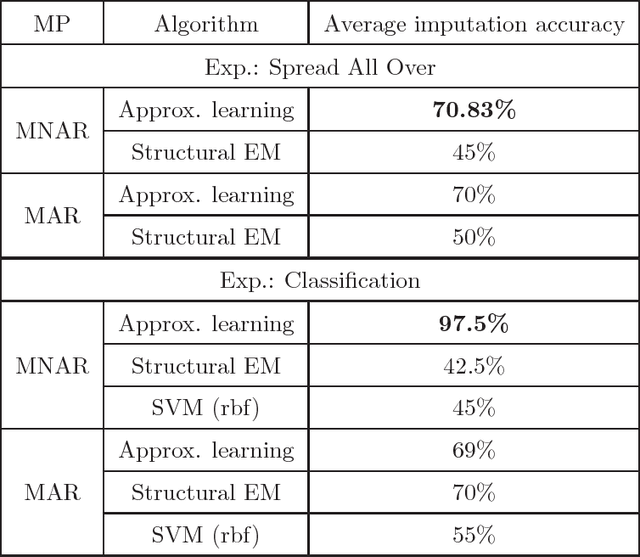
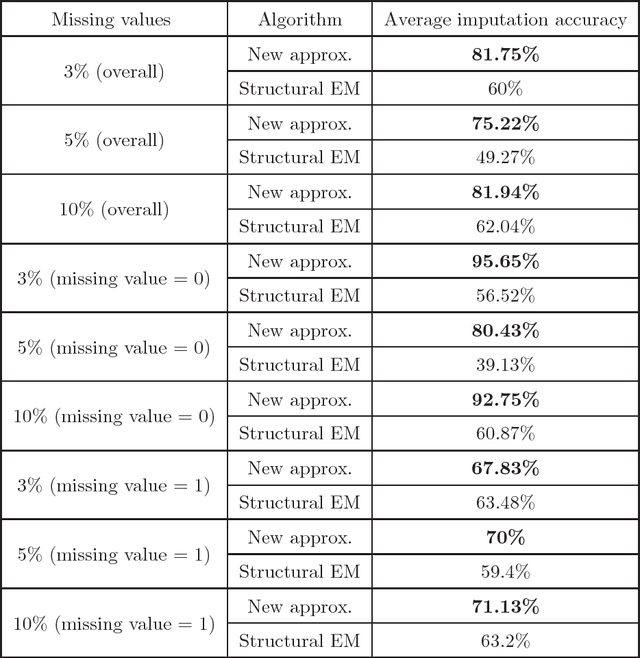
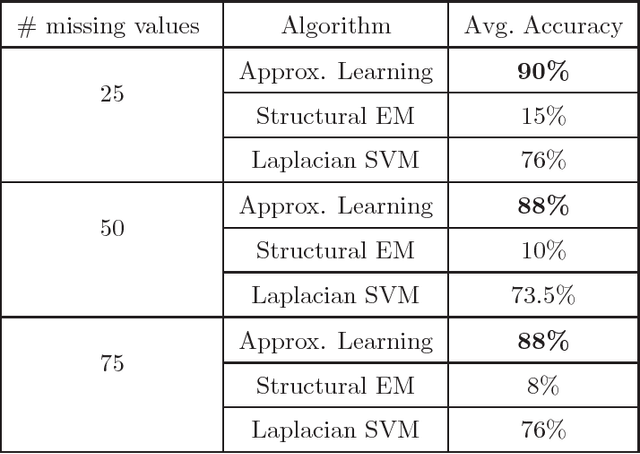
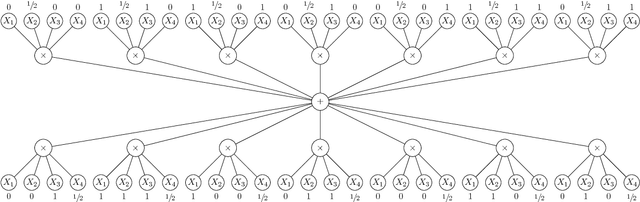
Abstract:We present new algorithms for learning Bayesian networks from data with missing values using a data augmentation approach. An exact Bayesian network learning algorithm is obtained by recasting the problem into a standard Bayesian network learning problem without missing data. To the best of our knowledge, this is the first exact algorithm for this problem. As expected, the exact algorithm does not scale to large domains. We build on the exact method to create an approximate algorithm using a hill-climbing technique. This algorithm scales to large domains so long as a suitable standard structure learning method for complete data is available. We perform a wide range of experiments to demonstrate the benefits of learning Bayesian networks with such new approach.
Learning Bounded Treewidth Bayesian Networks with Thousands of Variables
May 11, 2016



Abstract:We present a method for learning treewidth-bounded Bayesian networks from data sets containing thousands of variables. Bounding the treewidth of a Bayesian greatly reduces the complexity of inferences. Yet, being a global property of the graph, it considerably increases the difficulty of the learning process. We propose a novel algorithm for this task, able to scale to large domains and large treewidths. Our novel approach consistently outperforms the state of the art on data sets with up to ten thousand variables.
New Results for the MAP Problem in Bayesian Networks
Jul 29, 2010



Abstract:This paper presents new results for the (partial) maximum a posteriori (MAP) problem in Bayesian networks, which is the problem of querying the most probable state configuration of some of the network variables given evidence. First, it is demonstrated that the problem remains hard even in networks with very simple topology, such as binary polytrees and simple trees (including the Naive Bayes structure). Such proofs extend previous complexity results for the problem. Inapproximability results are also derived in the case of trees if the number of states per variable is not bounded. Although the problem is shown to be hard and inapproximable even in very simple scenarios, a new exact algorithm is described that is empirically fast in networks of bounded treewidth and bounded number of states per variable. The same algorithm is used as basis of a Fully Polynomial Time Approximation Scheme for MAP under such assumptions. Approximation schemes were generally thought to be impossible for this problem, but we show otherwise for classes of networks that are important in practice. The algorithms are extensively tested using some well-known networks as well as random generated cases to show their effectiveness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge