Brieuc Lehmann
Learning from data with structured missingness
Apr 04, 2023Abstract:Missing data are an unavoidable complication in many machine learning tasks. When data are `missing at random' there exist a range of tools and techniques to deal with the issue. However, as machine learning studies become more ambitious, and seek to learn from ever-larger volumes of heterogeneous data, an increasingly encountered problem arises in which missing values exhibit an association or structure, either explicitly or implicitly. Such `structured missingness' raises a range of challenges that have not yet been systematically addressed, and presents a fundamental hindrance to machine learning at scale. Here, we outline the current literature and propose a set of grand challenges in learning from data with structured missingness.
Density Estimation with Autoregressive Bayesian Predictives
Jun 13, 2022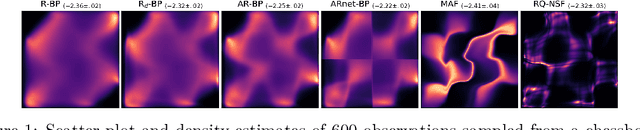
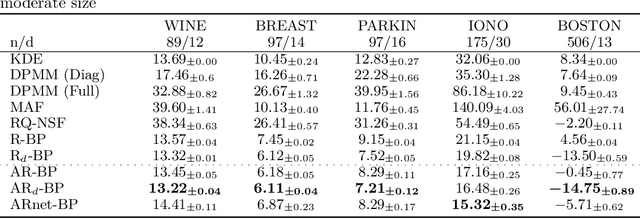
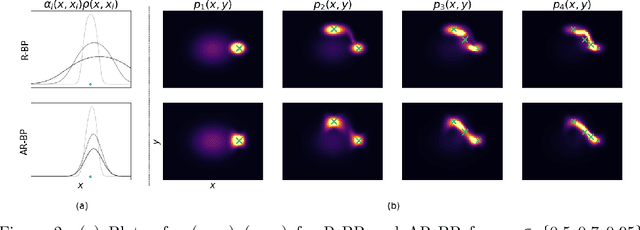
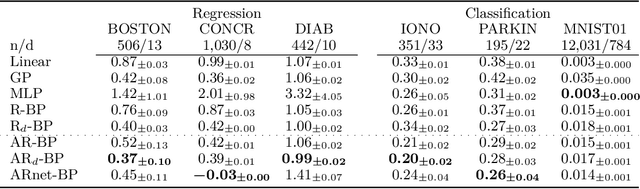
Abstract:Bayesian methods are a popular choice for statistical inference in small-data regimes due to the regularization effect induced by the prior, which serves to counteract overfitting. In the context of density estimation, the standard Bayesian approach is to target the posterior predictive. In general, direct estimation of the posterior predictive is intractable and so methods typically resort to approximating the posterior distribution as an intermediate step. The recent development of recursive predictive copula updates, however, has made it possible to perform tractable predictive density estimation without the need for posterior approximation. Although these estimators are computationally appealing, they tend to struggle on non-smooth data distributions. This is largely due to the comparatively restrictive form of the likelihood models from which the proposed copula updates were derived. To address this shortcoming, we consider a Bayesian nonparametric model with an autoregressive likelihood decomposition and Gaussian process prior, which yields a data-dependent bandwidth parameter in the copula update. Further, we formulate a novel parameterization of the bandwidth using an autoregressive neural network that maps the data into a latent space, and is thus able to capture more complex dependencies in the data. Our extensions increase the modelling capacity of existing recursive Bayesian density estimators, achieving state-of-the-art results on tabular data sets.
Neural Score Matching for High-Dimensional Causal Inference
Mar 01, 2022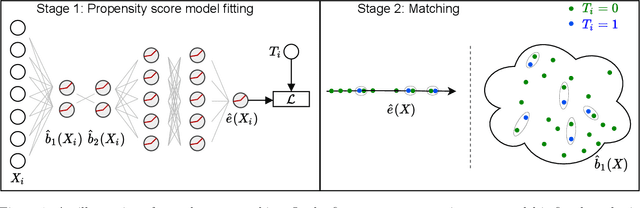
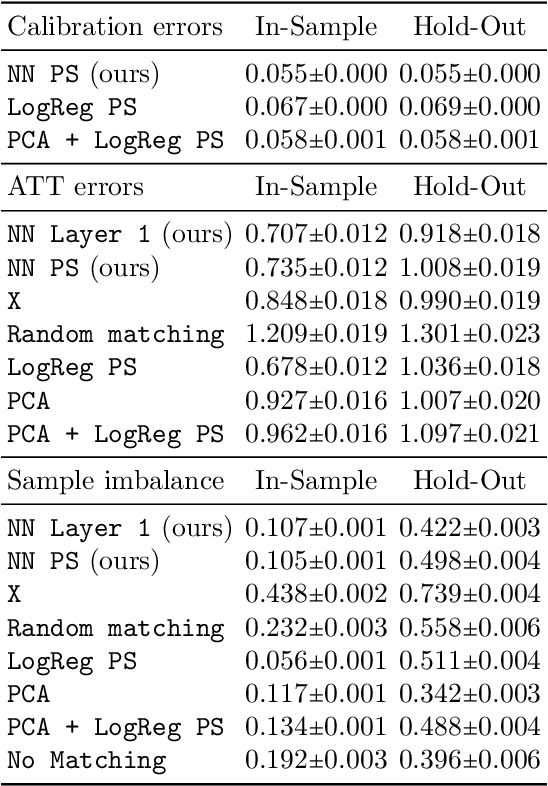


Abstract:Traditional methods for matching in causal inference are impractical for high-dimensional datasets. They suffer from the curse of dimensionality: exact matching and coarsened exact matching find exponentially fewer matches as the input dimension grows, and propensity score matching may match highly unrelated units together. To overcome this problem, we develop theoretical results which motivate the use of neural networks to obtain non-trivial, multivariate balancing scores of a chosen level of coarseness, in contrast to the classical, scalar propensity score. We leverage these balancing scores to perform matching for high-dimensional causal inference and call this procedure neural score matching. We show that our method is competitive against other matching approaches on semi-synthetic high-dimensional datasets, both in terms of treatment effect estimation and reducing imbalance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge