Ben Hoover
ConceptAttention: Diffusion Transformers Learn Highly Interpretable Features
Feb 06, 2025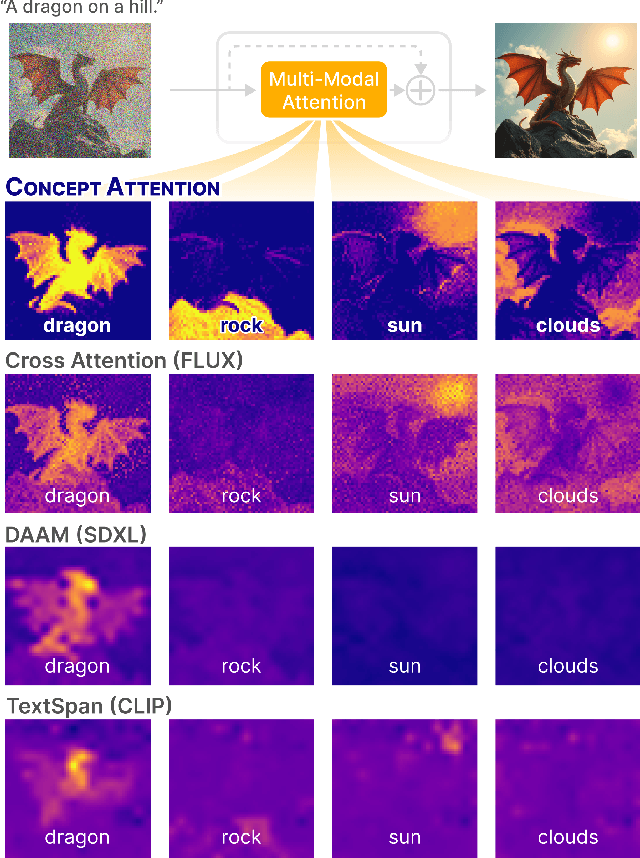
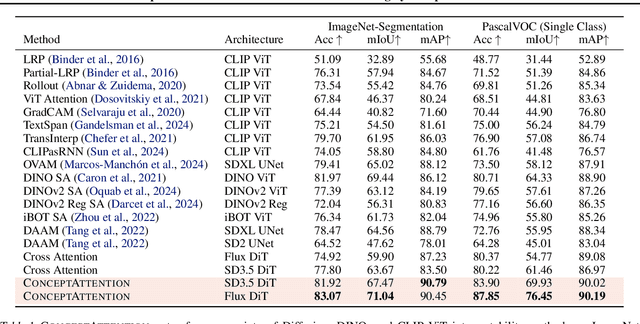
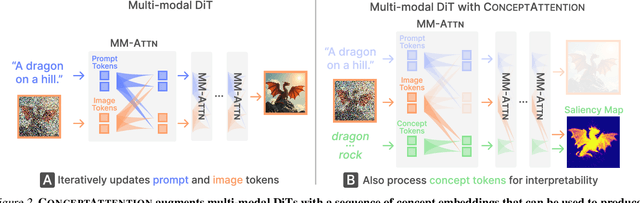
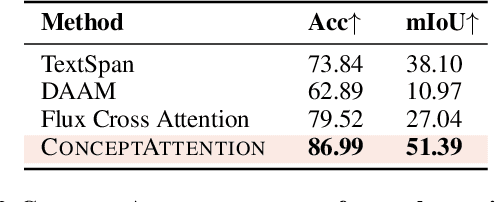
Abstract:Do the rich representations of multi-modal diffusion transformers (DiTs) exhibit unique properties that enhance their interpretability? We introduce ConceptAttention, a novel method that leverages the expressive power of DiT attention layers to generate high-quality saliency maps that precisely locate textual concepts within images. Without requiring additional training, ConceptAttention repurposes the parameters of DiT attention layers to produce highly contextualized concept embeddings, contributing the major discovery that performing linear projections in the output space of DiT attention layers yields significantly sharper saliency maps compared to commonly used cross-attention mechanisms. Remarkably, ConceptAttention even achieves state-of-the-art performance on zero-shot image segmentation benchmarks, outperforming 11 other zero-shot interpretability methods on the ImageNet-Segmentation dataset and on a single-class subset of PascalVOC. Our work contributes the first evidence that the representations of multi-modal DiT models like Flux are highly transferable to vision tasks like segmentation, even outperforming multi-modal foundation models like CLIP.
Target-Specific and Selective Drug Design for COVID-19 Using Deep Generative Models
Apr 02, 2020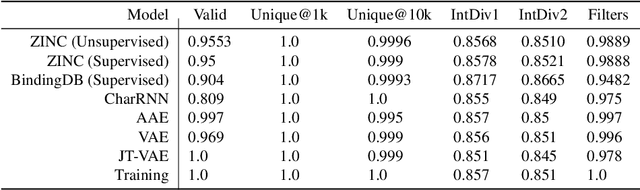
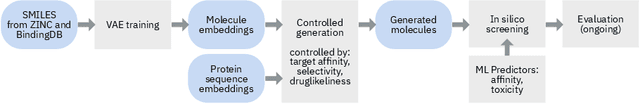
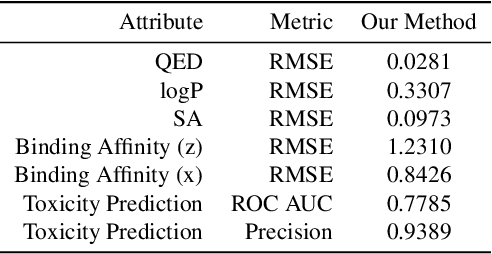
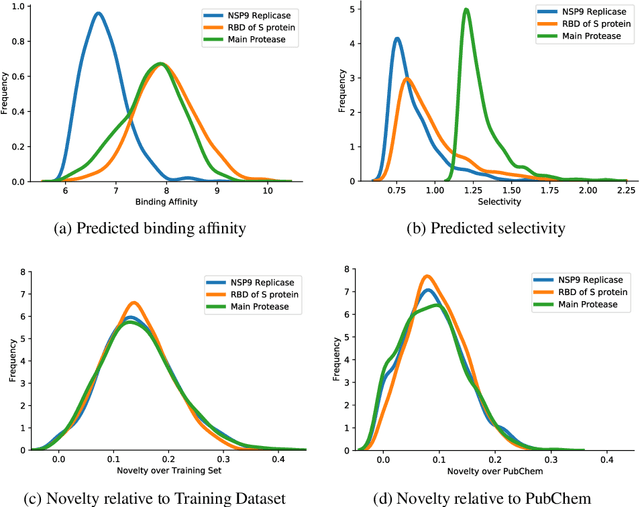
Abstract:The recent COVID-19 pandemic has highlighted the need for rapid therapeutic development for infectious diseases. To accelerate this process, we present a deep learning based generative modeling framework, CogMol, to design drug candidates specific to a given target protein sequence with high off-target selectivity. We augment this generative framework with an in silico screening process that accounts for toxicity, to lower the failure rate of the generated drug candidates in later stages of the drug development pipeline. We apply this framework to three relevant proteins of the SARS-CoV-2, the virus responsible for COVID-19, namely non-structural protein 9 (NSP9) replicase, main protease, and the receptor-binding domain (RBD) of the S protein. Docking to the target proteins demonstrate the potential of these generated molecules as ligands. Structural similarity analyses further imply novelty of the generated molecules with respect to the training dataset as well as possible biological association of a number of generated molecules that might be of relevance to COVID-19 therapeutic design. While the validation of these molecules is underway, we release ~ 3000 novel COVID-19 drug candidates generated using our framework. URL : http://ibm.biz/covid19-mol
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge