Barbara Pernici
Information Retrieval for Climate Impact
Apr 01, 2025Abstract:The purpose of the MANILA24 Workshop on information retrieval for climate impact was to bring together researchers from academia, industry, governments, and NGOs to identify and discuss core research problems in information retrieval to assess climate change impacts. The workshop aimed to foster collaboration by bringing communities together that have so far not been very well connected -- information retrieval, natural language processing, systematic reviews, impact assessments, and climate science. The workshop brought together a diverse set of researchers and practitioners interested in contributing to the development of a technical research agenda for information retrieval to assess climate change impacts.
Analyzing social media with crowdsourcing in Crowd4SDG
Aug 04, 2022


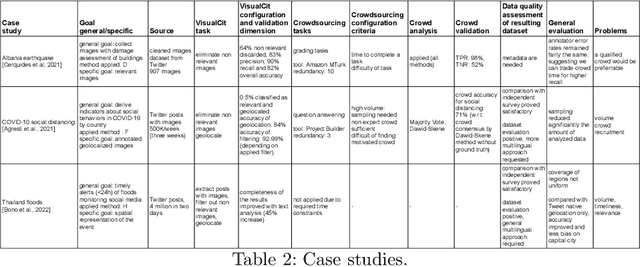
Abstract:Social media have the potential to provide timely information about emergency situations and sudden events. However, finding relevant information among millions of posts being posted every day can be difficult, and developing a data analysis project usually requires time and technical skills. This study presents an approach that provides flexible support for analyzing social media, particularly during emergencies. Different use cases in which social media analysis can be adopted are introduced, and the challenges of retrieving information from large sets of posts are discussed. The focus is on analyzing images and text contained in social media posts and a set of automatic data processing tools for filtering, classification, and geolocation of content with a human-in-the-loop approach to support the data analyst. Such support includes both feedback and suggestions to configure automated tools, and crowdsourcing to gather inputs from citizens. The results are validated by discussing three case studies developed within the Crowd4SDG H2020 European project.
Evaluating Scalable Uncertainty Estimation Methods for DNN-Based Molecular Property Prediction
Oct 07, 2019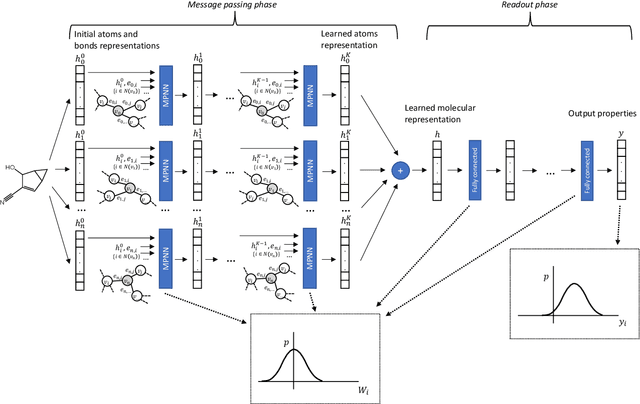

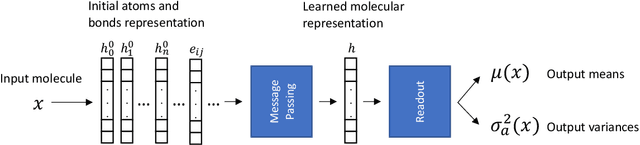
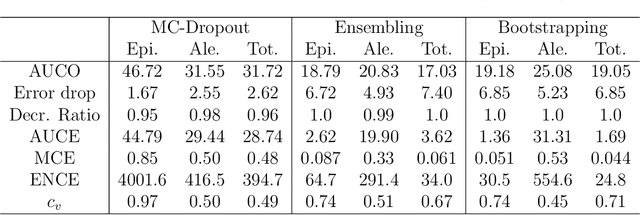
Abstract:Advances in deep neural network (DNN) based molecular property prediction have recently led to the development of models of remarkable accuracy and generalization ability, with graph convolution neural networks (GCNNs) reporting state-of-the-art performance for this task. However, some challenges remain and one of the most important that needs to be fully addressed concerns uncertainty quantification. DNN performance is affected by the volume and the quality of the training samples. Therefore, establishing when and to what extent a prediction can be considered reliable is just as important as outputting accurate predictions, especially when out-of-domain molecules are targeted. Recently, several methods to account for uncertainty in DNNs have been proposed, most of which are based on approximate Bayesian inference. Among these, only a few scale to the large datasets required in applications. Evaluating and comparing these methods has recently attracted great interest, but results are generally fragmented and absent for molecular property prediction. In this paper, we aim to quantitatively compare scalable techniques for uncertainty estimation in GCNNs. We introduce a set of quantitative criteria to capture different uncertainty aspects, and then use these criteria to compare MC-Dropout, deep ensembles, and bootstrapping, both theoretically in a unified framework that separates aleatoric/epistemic uncertainty and experimentally on the QM9 dataset. Our experiments quantify the performance of the different uncertainty estimation methods and their impact on uncertainty-related error reduction. Our findings indicate that ensembling and bootstrapping consistently outperform MC-Dropout, with different context-specific pros and cons. Our analysis also leads to a better understanding of the role of aleatoric/epistemic uncertainty and highlights the challenge posed by out-of-domain uncertainty.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge