Ann B. Ragin
Community-preserving Graph Convolutions for Structural and Functional Joint Embedding of Brain Networks
Nov 08, 2019



Abstract:Brain networks have received considerable attention given the critical significance for understanding human brain organization, for investigating neurological disorders and for clinical diagnostic applications. Structural brain network (e.g. DTI) and functional brain network (e.g. fMRI) are the primary networks of interest. Most existing works in brain network analysis focus on either structural or functional connectivity, which cannot leverage the complementary information from each other. Although multi-view learning methods have been proposed to learn from both networks (or views), these methods aim to reach a consensus among multiple views, and thus distinct intrinsic properties of each view may be ignored. How to jointly learn representations from structural and functional brain networks while preserving their inherent properties is a critical problem. In this paper, we propose a framework of Siamese community-preserving graph convolutional network (SCP-GCN) to learn the structural and functional joint embedding of brain networks. Specifically, we use graph convolutions to learn the structural and functional joint embedding, where the graph structure is defined with structural connectivity and node features are from the functional connectivity. Moreover, we propose to preserve the community structure of brain networks in the graph convolutions by considering the intra-community and inter-community properties in the learning process. Furthermore, we use Siamese architecture which models the pair-wise similarity learning to guide the learning process. To evaluate the proposed approach, we conduct extensive experiments on two real brain network datasets. The experimental results demonstrate the superior performance of the proposed approach in structural and functional joint embedding for neurological disorder analysis, indicating its promising value for clinical applications.
Multi-View Multi-Graph Embedding for Brain Network Clustering Analysis
Jun 19, 2018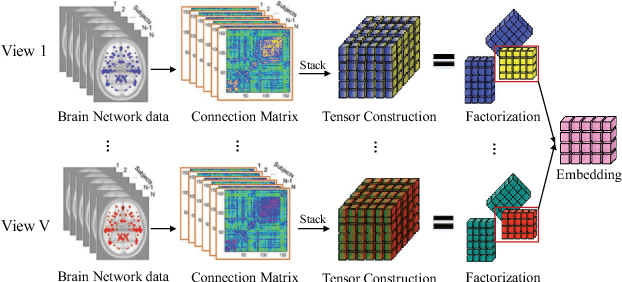

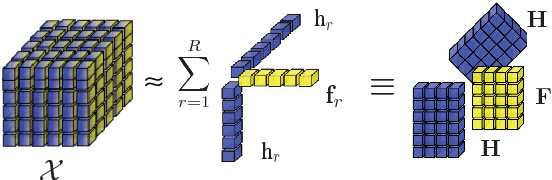
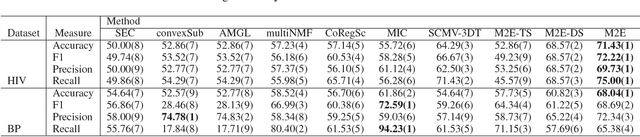
Abstract:Network analysis of human brain connectivity is critically important for understanding brain function and disease states. Embedding a brain network as a whole graph instance into a meaningful low-dimensional representation can be used to investigate disease mechanisms and inform therapeutic interventions. Moreover, by exploiting information from multiple neuroimaging modalities or views, we are able to obtain an embedding that is more useful than the embedding learned from an individual view. Therefore, multi-view multi-graph embedding becomes a crucial task. Currently, only a few studies have been devoted to this topic, and most of them focus on the vector-based strategy which will cause structural information contained in the original graphs lost. As a novel attempt to tackle this problem, we propose Multi-view Multi-graph Embedding (M2E) by stacking multi-graphs into multiple partially-symmetric tensors and using tensor techniques to simultaneously leverage the dependencies and correlations among multi-view and multi-graph brain networks. Extensive experiments on real HIV and bipolar disorder brain network datasets demonstrate the superior performance of M2E on clustering brain networks by leveraging the multi-view multi-graph interactions.
Multi-view Graph Embedding with Hub Detection for Brain Network Analysis
Sep 12, 2017


Abstract:Multi-view graph embedding has become a widely studied problem in the area of graph learning. Most of the existing works on multi-view graph embedding aim to find a shared common node embedding across all the views of the graph by combining the different views in a specific way. Hub detection, as another essential topic in graph mining has also drawn extensive attentions in recent years, especially in the context of brain network analysis. Both the graph embedding and hub detection relate to the node clustering structure of graphs. The multi-view graph embedding usually implies the node clustering structure of the graph based on the multiple views, while the hubs are the boundary-spanning nodes across different node clusters in the graph and thus may potentially influence the clustering structure of the graph. However, none of the existing works in multi-view graph embedding considered the hubs when learning the multi-view embeddings. In this paper, we propose to incorporate the hub detection task into the multi-view graph embedding framework so that the two tasks could benefit each other. Specifically, we propose an auto-weighted framework of Multi-view Graph Embedding with Hub Detection (MVGE-HD) for brain network analysis. The MVGE-HD framework learns a unified graph embedding across all the views while reducing the potential influence of the hubs on blurring the boundaries between node clusters in the graph, thus leading to a clear and discriminative node clustering structure for the graph. We apply MVGE-HD on two real multi-view brain network datasets (i.e., HIV and Bipolar). The experimental results demonstrate the superior performance of the proposed framework in brain network analysis for clinical investigation and application.
Mining Brain Networks using Multiple Side Views for Neurological Disorder Identification
Aug 19, 2015



Abstract:Mining discriminative subgraph patterns from graph data has attracted great interest in recent years. It has a wide variety of applications in disease diagnosis, neuroimaging, etc. Most research on subgraph mining focuses on the graph representation alone. However, in many real-world applications, the side information is available along with the graph data. For example, for neurological disorder identification, in addition to the brain networks derived from neuroimaging data, hundreds of clinical, immunologic, serologic and cognitive measures may also be documented for each subject. These measures compose multiple side views encoding a tremendous amount of supplemental information for diagnostic purposes, yet are often ignored. In this paper, we study the problem of discriminative subgraph selection using multiple side views and propose a novel solution to find an optimal set of subgraph features for graph classification by exploring a plurality of side views. We derive a feature evaluation criterion, named gSide, to estimate the usefulness of subgraph patterns based upon side views. Then we develop a branch-and-bound algorithm, called gMSV, to efficiently search for optimal subgraph features by integrating the subgraph mining process and the procedure of discriminative feature selection. Empirical studies on graph classification tasks for neurological disorders using brain networks demonstrate that subgraph patterns selected by the multi-side-view guided subgraph selection approach can effectively boost graph classification performances and are relevant to disease diagnosis.
DuSK: A Dual Structure-preserving Kernel for Supervised Tensor Learning with Applications to Neuroimages
Aug 05, 2014



Abstract:With advances in data collection technologies, tensor data is assuming increasing prominence in many applications and the problem of supervised tensor learning has emerged as a topic of critical significance in the data mining and machine learning community. Conventional methods for supervised tensor learning mainly focus on learning kernels by flattening the tensor into vectors or matrices, however structural information within the tensors will be lost. In this paper, we introduce a new scheme to design structure-preserving kernels for supervised tensor learning. Specifically, we demonstrate how to leverage the naturally available structure within the tensorial representation to encode prior knowledge in the kernel. We proposed a tensor kernel that can preserve tensor structures based upon dual-tensorial mapping. The dual-tensorial mapping function can map each tensor instance in the input space to another tensor in the feature space while preserving the tensorial structure. Theoretically, our approach is an extension of the conventional kernels in the vector space to tensor space. We applied our novel kernel in conjunction with SVM to real-world tensor classification problems including brain fMRI classification for three different diseases (i.e., Alzheimer's disease, ADHD and brain damage by HIV). Extensive empirical studies demonstrate that our proposed approach can effectively boost tensor classification performances, particularly with small sample sizes.
Discriminative Feature Selection for Uncertain Graph Classification
Jan 28, 2013



Abstract:Mining discriminative features for graph data has attracted much attention in recent years due to its important role in constructing graph classifiers, generating graph indices, etc. Most measurement of interestingness of discriminative subgraph features are defined on certain graphs, where the structure of graph objects are certain, and the binary edges within each graph represent the "presence" of linkages among the nodes. In many real-world applications, however, the linkage structure of the graphs is inherently uncertain. Therefore, existing measurements of interestingness based upon certain graphs are unable to capture the structural uncertainty in these applications effectively. In this paper, we study the problem of discriminative subgraph feature selection from uncertain graphs. This problem is challenging and different from conventional subgraph mining problems because both the structure of the graph objects and the discrimination score of each subgraph feature are uncertain. To address these challenges, we propose a novel discriminative subgraph feature selection method, DUG, which can find discriminative subgraph features in uncertain graphs based upon different statistical measures including expectation, median, mode and phi-probability. We first compute the probability distribution of the discrimination scores for each subgraph feature based on dynamic programming. Then a branch-and-bound algorithm is proposed to search for discriminative subgraphs efficiently. Extensive experiments on various neuroimaging applications (i.e., Alzheimer's Disease, ADHD and HIV) have been performed to analyze the gain in performance by taking into account structural uncertainties in identifying discriminative subgraph features for graph classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge