Andrew White
Graph AI generates neurological hypotheses validated in molecular, organoid, and clinical systems
Dec 13, 2025Abstract:Neurological diseases are the leading global cause of disability, yet most lack disease-modifying treatments. We present PROTON, a heterogeneous graph transformer that generates testable hypotheses across molecular, organoid, and clinical systems. To evaluate PROTON, we apply it to Parkinson's disease (PD), bipolar disorder (BD), and Alzheimer's disease (AD). In PD, PROTON linked genetic risk loci to genes essential for dopaminergic neuron survival and predicted pesticides toxic to patient-derived neurons, including the insecticide endosulfan, which ranked within the top 1.29% of predictions. In silico screens performed by PROTON reproduced six genome-wide $α$-synuclein experiments, including a split-ubiquitin yeast two-hybrid system (normalized enrichment score [NES] = 2.30, FDR-adjusted $p < 1 \times 10^{-4}$), an ascorbate peroxidase proximity labeling assay (NES = 2.16, FDR $< 1 \times 10^{-4}$), and a high-depth targeted exome sequencing study in 496 synucleinopathy patients (NES = 2.13, FDR $< 1 \times 10^{-4}$). In BD, PROTON predicted calcitriol as a candidate drug that reversed proteomic alterations observed in cortical organoids derived from BD patients. In AD, we evaluated PROTON predictions in health records from $n = 610,524$ patients at Mass General Brigham, confirming that five PROTON-predicted drugs were associated with reduced seven-year dementia risk (minimum hazard ratio = 0.63, 95% CI: 0.53-0.75, $p < 1 \times 10^{-7}$). PROTON generated neurological hypotheses that were evaluated across molecular, organoid, and clinical systems, defining a path for AI-driven discovery in neurological disease.
SELFIES and the future of molecular string representations
Mar 31, 2022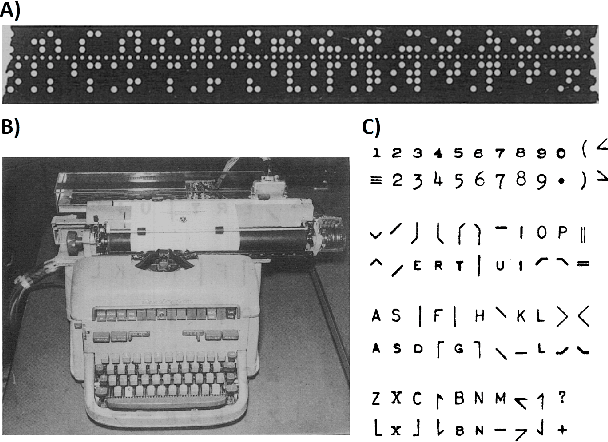
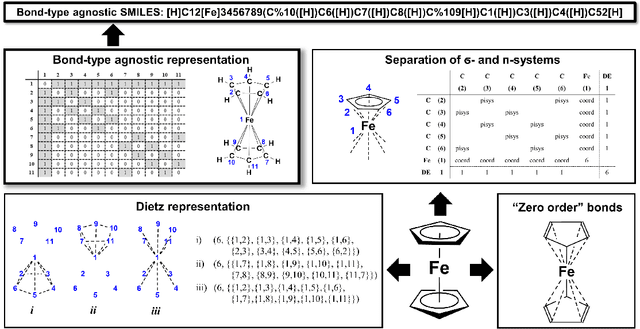
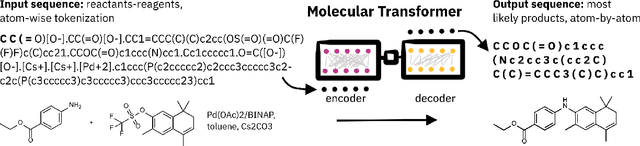
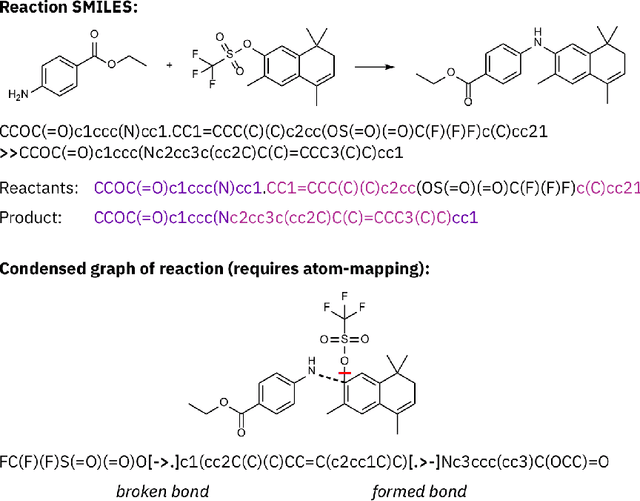
Abstract:Artificial intelligence (AI) and machine learning (ML) are expanding in popularity for broad applications to challenging tasks in chemistry and materials science. Examples include the prediction of properties, the discovery of new reaction pathways, or the design of new molecules. The machine needs to read and write fluently in a chemical language for each of these tasks. Strings are a common tool to represent molecular graphs, and the most popular molecular string representation, SMILES, has powered cheminformatics since the late 1980s. However, in the context of AI and ML in chemistry, SMILES has several shortcomings -- most pertinently, most combinations of symbols lead to invalid results with no valid chemical interpretation. To overcome this issue, a new language for molecules was introduced in 2020 that guarantees 100\% robustness: SELFIES (SELF-referencIng Embedded Strings). SELFIES has since simplified and enabled numerous new applications in chemistry. In this manuscript, we look to the future and discuss molecular string representations, along with their respective opportunities and challenges. We propose 16 concrete Future Projects for robust molecular representations. These involve the extension toward new chemical domains, exciting questions at the interface of AI and robust languages and interpretability for both humans and machines. We hope that these proposals will inspire several follow-up works exploiting the full potential of molecular string representations for the future of AI in chemistry and materials science.
Federated Learning of Molecular Properties in a Heterogeneous Setting
Sep 15, 2021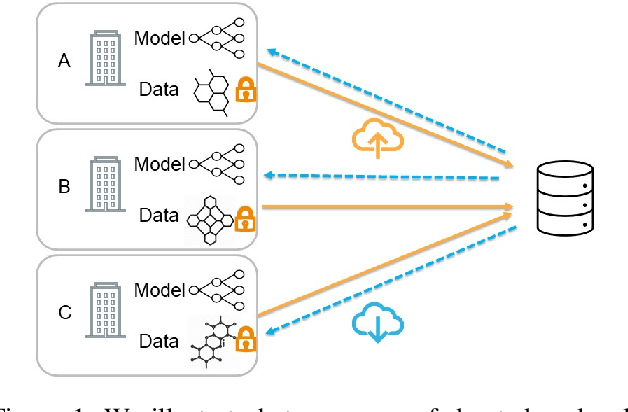
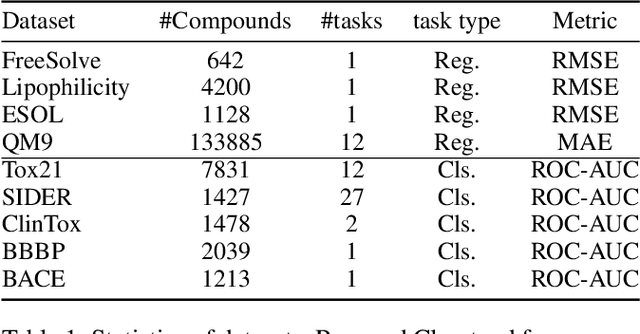
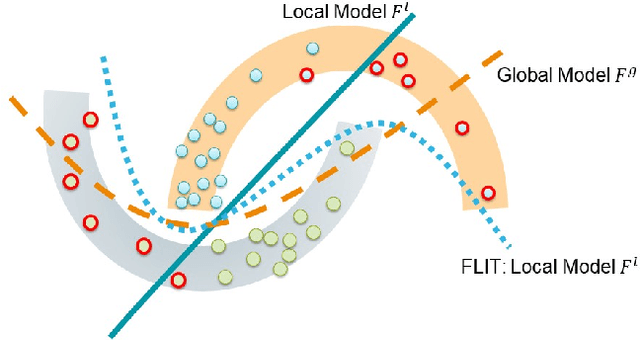
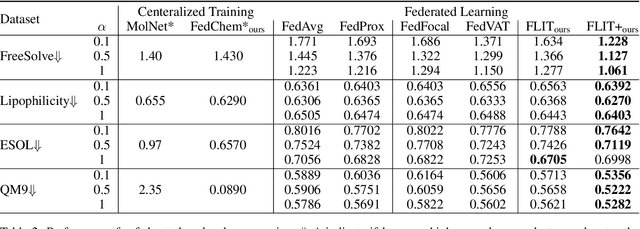
Abstract:Chemistry research has both high material and computational costs to conduct experiments. Institutions thus consider chemical data to be valuable and there have been few efforts to construct large public datasets for machine learning. Another challenge is that different intuitions are interested in different classes of molecules, creating heterogeneous data that cannot be easily joined by conventional distributed training. In this work, we introduce federated heterogeneous molecular learning to address these challenges. Federated learning allows end-users to build a global model collaboratively while preserving the training data distributed over isolated clients. Due to the lack of related research, we first simulate a federated heterogeneous benchmark called FedChem. FedChem is constructed by jointly performing scaffold splitting and Latent Dirichlet Allocation on existing datasets. Our results on FedChem show that significant learning challenges arise when working with heterogeneous molecules. We then propose a method to alleviate the problem, namely Federated Learning by Instance reweighTing (FLIT). FLIT can align the local training across heterogeneous clients by improving the performance for uncertain samples. Comprehensive experiments conducted on our new benchmark FedChem validate the advantages of this method over other federated learning schemes. FedChem should enable a new type of collaboration for improving AI in chemistry that mitigates concerns about valuable chemical data.
An Easy to Use Repository for Comparing and Improving Machine Learning Algorithm Usage
Jun 05, 2014
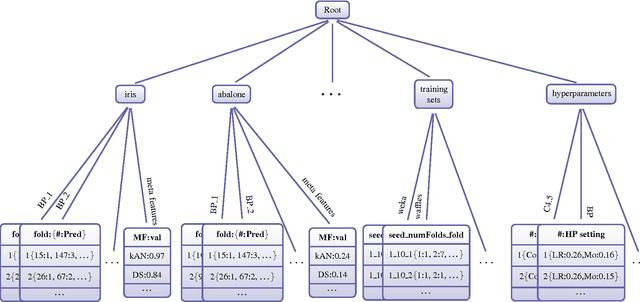
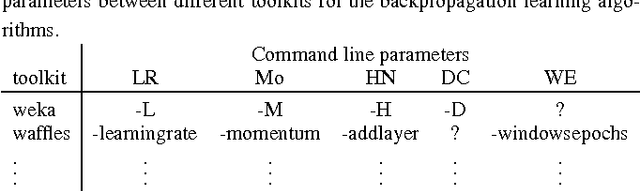
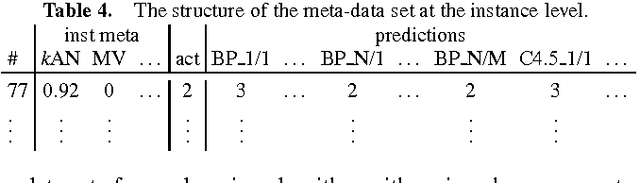
Abstract:The results from most machine learning experiments are used for a specific purpose and then discarded. This results in a significant loss of information and requires rerunning experiments to compare learning algorithms. This also requires implementation of another algorithm for comparison, that may not always be correctly implemented. By storing the results from previous experiments, machine learning algorithms can be compared easily and the knowledge gained from them can be used to improve their performance. The purpose of this work is to provide easy access to previous experimental results for learning and comparison. These stored results are comprehensive -- storing the prediction for each test instance as well as the learning algorithm, hyperparameters, and training set that were used. Previous results are particularly important for meta-learning, which, in a broad sense, is the process of learning from previous machine learning results such that the learning process is improved. While other experiment databases do exist, one of our focuses is on easy access to the data. We provide meta-learning data sets that are ready to be downloaded for meta-learning experiments. In addition, queries to the underlying database can be made if specific information is desired. We also differ from previous experiment databases in that our databases is designed at the instance level, where an instance is an example in a data set. We store the predictions of a learning algorithm trained on a specific training set for each instance in the test set. Data set level information can then be obtained by aggregating the results from the instances. The instance level information can be used for many tasks such as determining the diversity of a classifier or algorithmically determining the optimal subset of training instances for a learning algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge