Amy Kuceyeski
Decoding natural image stimuli from fMRI data with a surface-based convolutional network
Dec 05, 2022Abstract:Due to the low signal-to-noise ratio and limited resolution of functional MRI data, and the high complexity of natural images, reconstructing a visual stimulus from human brain fMRI measurements is a challenging task. In this work, we propose a novel approach for this task, which we call Cortex2Image, to decode visual stimuli with high semantic fidelity and rich fine-grained detail. In particular, we train a surface-based convolutional network model that maps from brain response to semantic image features first (Cortex2Semantic). We then combine this model with a high-quality image generator (Instance-Conditioned GAN) to train another mapping from brain response to fine-grained image features using a variational approach (Cortex2Detail). Image reconstructions obtained by our proposed method achieve state-of-the-art semantic fidelity, while yielding good fine-grained similarity with the ground-truth stimulus. Our code is available at: https://github.com/zijin-gu/meshconv-decoding.git.
Personalized visual encoding model construction with small data
Feb 04, 2022
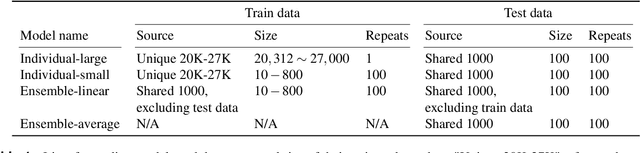
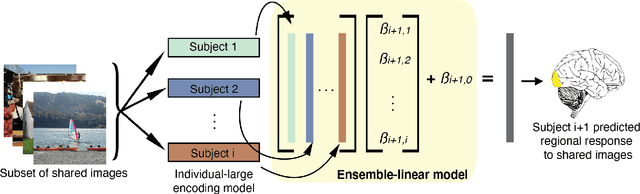
Abstract:Encoding models that predict brain response patterns to stimuli are one way to capture this relationship between variability in bottom-up neural systems and individual's behavior or pathological state. However, they generally need a large amount of training data to achieve optimal accuracy. Here, we propose and test an alternative personalized ensemble encoding model approach to utilize existing encoding models, to create encoding models for novel individuals with relatively little stimuli-response data. We show that these personalized ensemble encoding models trained with small amounts of data for a specific individual, i.e. ~400 image-response pairs, achieve accuracy not different from models trained on ~24,000 image-response pairs for the same individual. Importantly, the personalized ensemble encoding models preserve patterns of inter-individual variability in the image-response relationship. Additionally, we use our personalized ensemble encoding model within the recently developed NeuroGen framework to generate optimal stimuli designed to maximize specific regions' activations for a specific individual. We show that the inter-individual differences in face area responses to images of dog vs human faces observed previously is replicated using NeuroGen with the ensemble encoding model. Finally, and most importantly, we show the proposed approach is robust against domain shift by validating on a prospectively collected set of image-response data in novel individuals with a different scanner and experimental setup. Our approach shows the potential to use previously collected, deeply sampled data to efficiently create accurate, personalized encoding models and, subsequently, personalized optimal synthetic images for new individuals scanned under different experimental conditions.
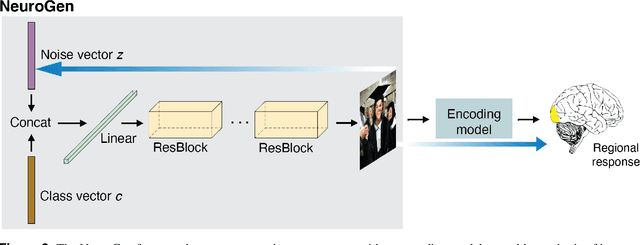
NeuroGen: activation optimized image synthesis for discovery neuroscience
May 15, 2021



Abstract:Functional MRI (fMRI) is a powerful technique that has allowed us to characterize visual cortex responses to stimuli, yet such experiments are by nature constructed based on a priori hypotheses, limited to the set of images presented to the individual while they are in the scanner, are subject to noise in the observed brain responses, and may vary widely across individuals. In this work, we propose a novel computational strategy, which we call NeuroGen, to overcome these limitations and develop a powerful tool for human vision neuroscience discovery. NeuroGen combines an fMRI-trained neural encoding model of human vision with a deep generative network to synthesize images predicted to achieve a target pattern of macro-scale brain activation. We demonstrate that the reduction of noise that the encoding model provides, coupled with the generative network's ability to produce images of high fidelity, results in a robust discovery architecture for visual neuroscience. By using only a small number of synthetic images created by NeuroGen, we demonstrate that we can detect and amplify differences in regional and individual human brain response patterns to visual stimuli. We then verify that these discoveries are reflected in the several thousand observed image responses measured with fMRI. We further demonstrate that NeuroGen can create synthetic images predicted to achieve regional response patterns not achievable by the best-matching natural images. The NeuroGen framework extends the utility of brain encoding models and opens up a new avenue for exploring, and possibly precisely controlling, the human visual system.
Neural encoding with visual attention
Oct 01, 2020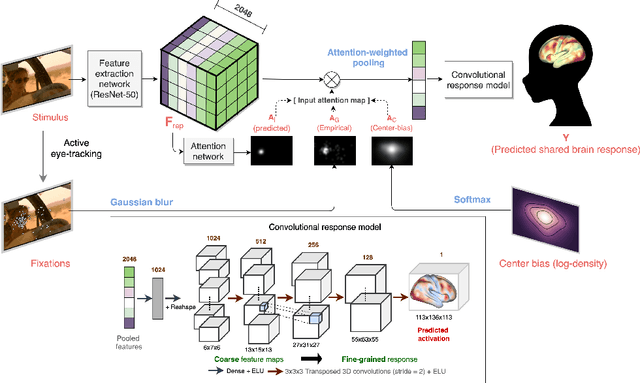
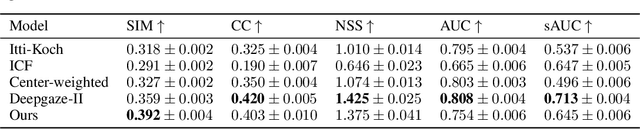
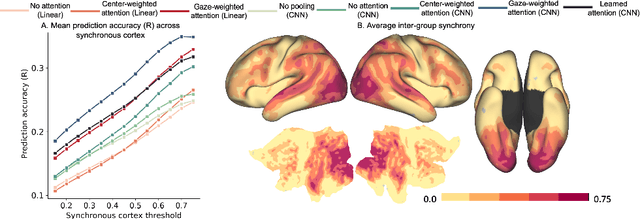
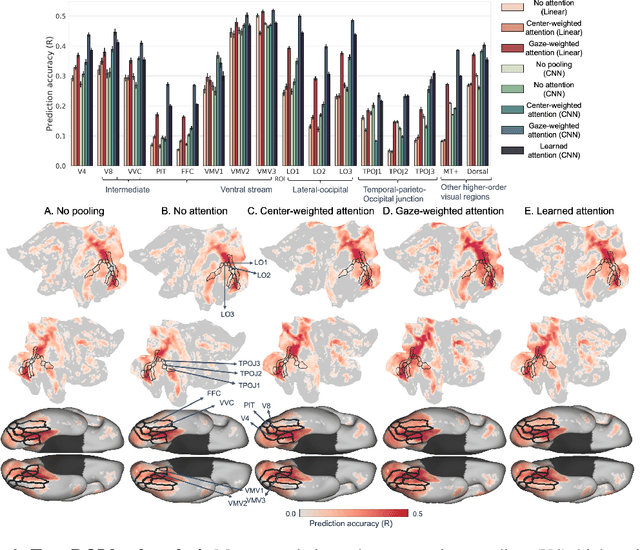
Abstract:Visual perception is critically influenced by the focus of attention. Due to limited resources, it is well known that neural representations are biased in favor of attended locations. Using concurrent eye-tracking and functional Magnetic Resonance Imaging (fMRI) recordings from a large cohort of human subjects watching movies, we first demonstrate that leveraging gaze information, in the form of attentional masking, can significantly improve brain response prediction accuracy in a neural encoding model. Next, we propose a novel approach to neural encoding by including a trainable soft-attention module. Using our new approach, we demonstrate that it is possible to learn visual attention policies by end-to-end learning merely on fMRI response data, and without relying on any eye-tracking. Interestingly, we find that attention locations estimated by the model on independent data agree well with the corresponding eye fixation patterns, despite no explicit supervision to do so. Together, these findings suggest that attention modules can be instrumental in neural encoding models of visual stimuli.
From Connectomic to Task-evoked Fingerprints: Individualized Prediction of Task Contrasts from Resting-state Functional Connectivity
Aug 07, 2020
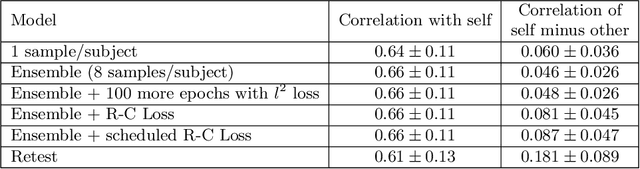


Abstract:Resting-state functional MRI (rsfMRI) yields functional connectomes that can serve as cognitive fingerprints of individuals. Connectomic fingerprints have proven useful in many machine learning tasks, such as predicting subject-specific behavioral traits or task-evoked activity. In this work, we propose a surface-based convolutional neural network (BrainSurfCNN) model to predict individual task contrasts from their resting-state fingerprints. We introduce a reconstructive-contrastive loss that enforces subject-specificity of model outputs while minimizing predictive error. The proposed approach significantly improves the accuracy of predicted contrasts over a well-established baseline. Furthermore, BrainSurfCNN's prediction also surpasses test-retest benchmark in a subject identification task.
A shared neural encoding model for the prediction of subject-specific fMRI response
Jul 11, 2020


Abstract:The increasing popularity of naturalistic paradigms in fMRI (such as movie watching) demands novel strategies for multi-subject data analysis, such as use of neural encoding models. In the present study, we propose a shared convolutional neural encoding method that accounts for individual-level differences. Our method leverages multi-subject data to improve the prediction of subject-specific responses evoked by visual or auditory stimuli. We showcase our approach on high-resolution 7T fMRI data from the Human Connectome Project movie-watching protocol and demonstrate significant improvement over single-subject encoding models. We further demonstrate the ability of the shared encoding model to successfully capture meaningful individual differences in response to traditional task-based facial and scenes stimuli. Taken together, our findings suggest that inter-subject knowledge transfer can be beneficial to subject-specific predictive models.
Detecting abnormalities in resting-state dynamics: An unsupervised learning approach
Aug 16, 2019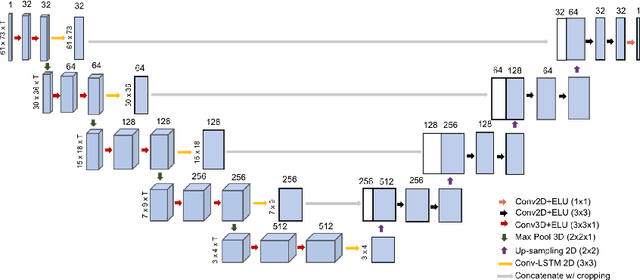
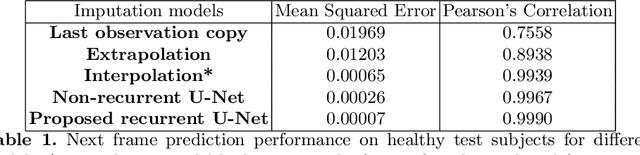
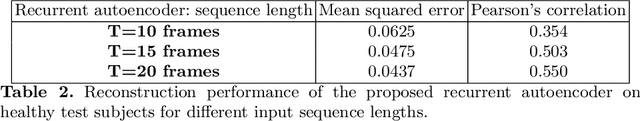
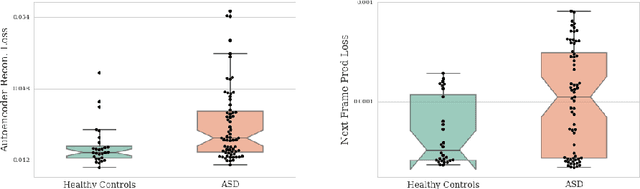
Abstract:Resting-state functional MRI (rs-fMRI) is a rich imaging modality that captures spontaneous brain activity patterns, revealing clues about the connectomic organization of the human brain. While many rs-fMRI studies have focused on static measures of functional connectivity, there has been a recent surge in examining the temporal patterns in these data. In this paper, we explore two strategies for capturing the normal variability in resting-state activity across a healthy population: (a) an autoencoder approach on the rs-fMRI sequence, and (b) a next frame prediction strategy. We show that both approaches can learn useful representations of rs-fMRI data and demonstrate their novel application for abnormality detection in the context of discriminating autism patients from healthy controls.
Machine learning in resting-state fMRI analysis
Dec 30, 2018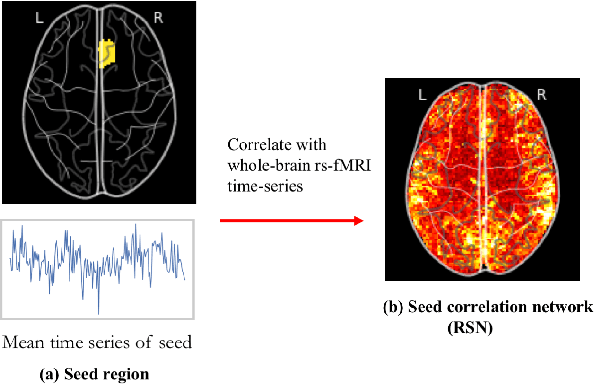
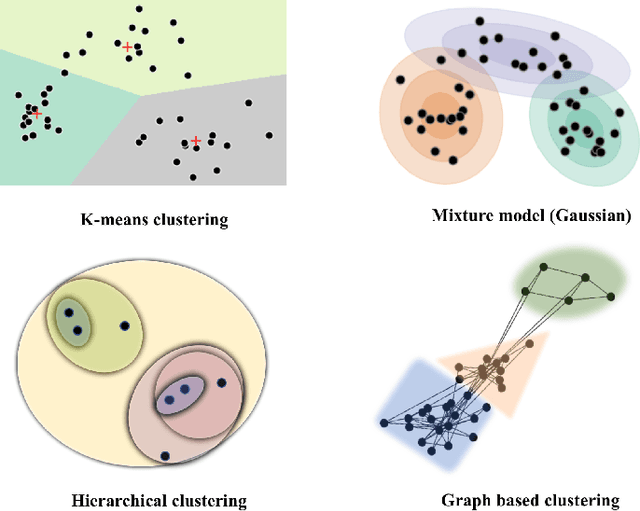
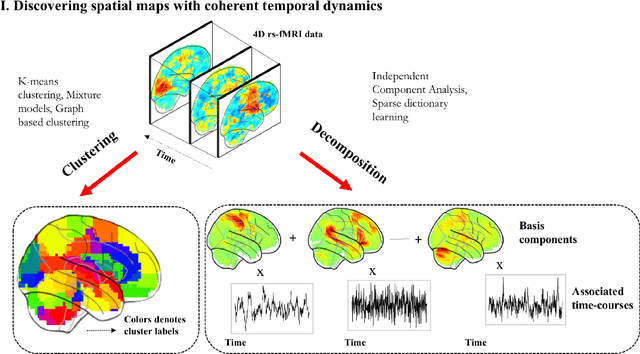
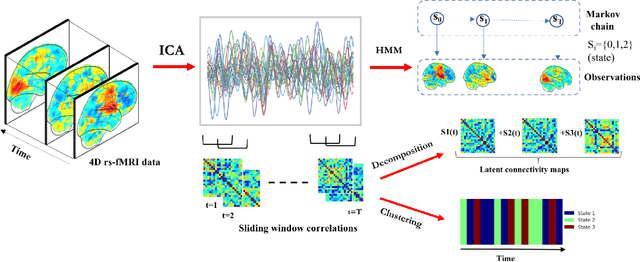
Abstract:Machine learning techniques have gained prominence for the analysis of resting-state functional Magnetic Resonance Imaging (rs-fMRI) data. Here, we present an overview of various unsupervised and supervised machine learning applications to rs-fMRI. We present a methodical taxonomy of machine learning methods in resting-state fMRI. We identify three major divisions of unsupervised learning methods with regard to their applications to rs-fMRI, based on whether they discover principal modes of variation across space, time or population. Next, we survey the algorithms and rs-fMRI feature representations that have driven the success of supervised subject-level predictions. The goal is to provide a high-level overview of the burgeoning field of rs-fMRI from the perspective of machine learning applications.
Large Spectral Density Matrix Estimation by Thresholding
Dec 03, 2018



Abstract:Spectral density matrix estimation of multivariate time series is a classical problem in time series and signal processing. In modern neuroscience, spectral density based metrics are commonly used for analyzing functional connectivity among brain regions. In this paper, we develop a non-asymptotic theory for regularized estimation of high-dimensional spectral density matrices of Gaussian and linear processes using thresholded versions of averaged periodograms. Our theoretical analysis ensures that consistent estimation of spectral density matrix of a $p$-dimensional time series using $n$ samples is possible under high-dimensional regime $\log p / n \rightarrow 0$ as long as the true spectral density is approximately sparse. A key technical component of our analysis is a new concentration inequality of average periodogram around its expectation, which is of independent interest. Our estimation consistency results complement existing results for shrinkage based estimators of multivariate spectral density, which require no assumption on sparsity but only ensure consistent estimation in a regime $p^2/n \rightarrow 0$. In addition, our proposed thresholding based estimators perform consistent and automatic edge selection when learning coherence networks among the components of a multivariate time series. We demonstrate the advantage of our estimators using simulation studies and a real data application on functional connectivity analysis with fMRI data.
Ensemble learning with 3D convolutional neural networks for connectome-based prediction
Sep 11, 2018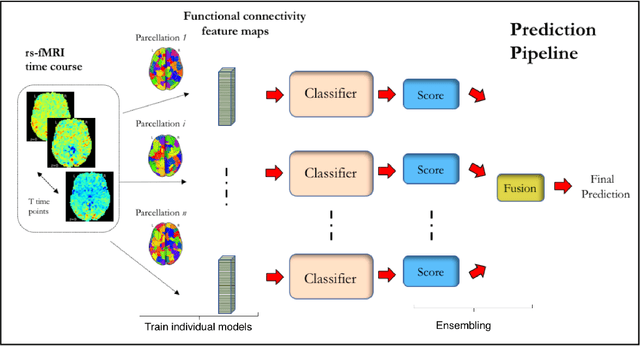
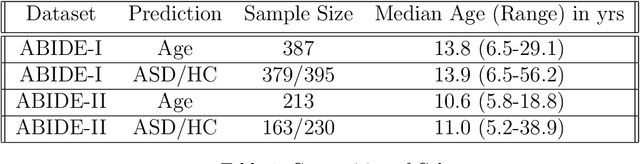
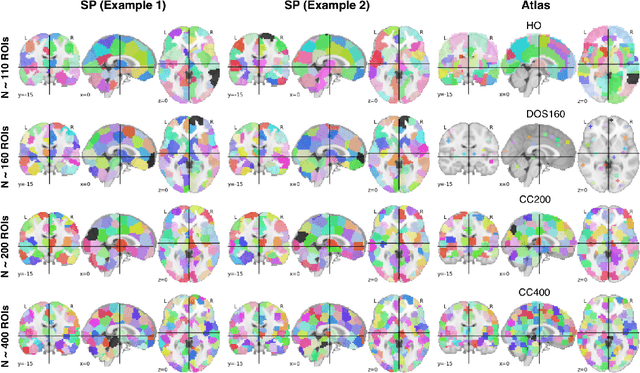
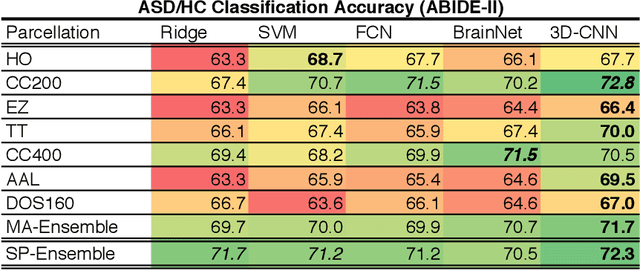
Abstract:The specificty and sensitivity of resting state functional MRI (rs-fMRI) measurements depend on pre-processing choices, such as the parcellation scheme used to define regions of interest (ROIs). In this study, we critically evaluate the effect of brain parcellations on machine learning models applied to rs-fMRI data. Our experiments reveal a remarkable trend: On average, models with stochastic parcellations consistently perform as well as models with widely used atlases at the same spatial scale. We thus propose an ensemble learning strategy to combine the predictions from models trained on connectivity data extracted using different (e.g., stochastic) parcellations. We further present an implementation of our ensemble learning strategy with a novel 3D Convolutional Neural Network (CNN) approach. The proposed CNN approach takes advantage of the full-resolution 3D spatial structure of rs-fMRI data and fits non-linear predictive models. Our ensemble CNN framework overcomes the limitations of traditional machine learning models for connectomes that often rely on region-based summary statistics and/or linear models. We showcase our approach on a classification (autism patients versus healthy controls) and a regression problem (prediction of subject's age), and report promising results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge