Amir Ghasemian
Stacking Models for Nearly Optimal Link Prediction in Complex Networks
Sep 17, 2019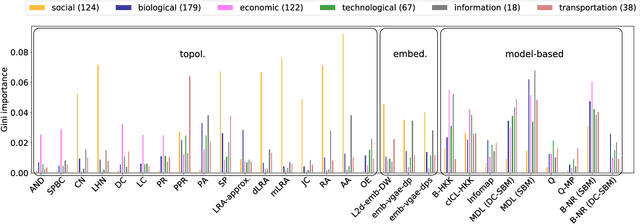
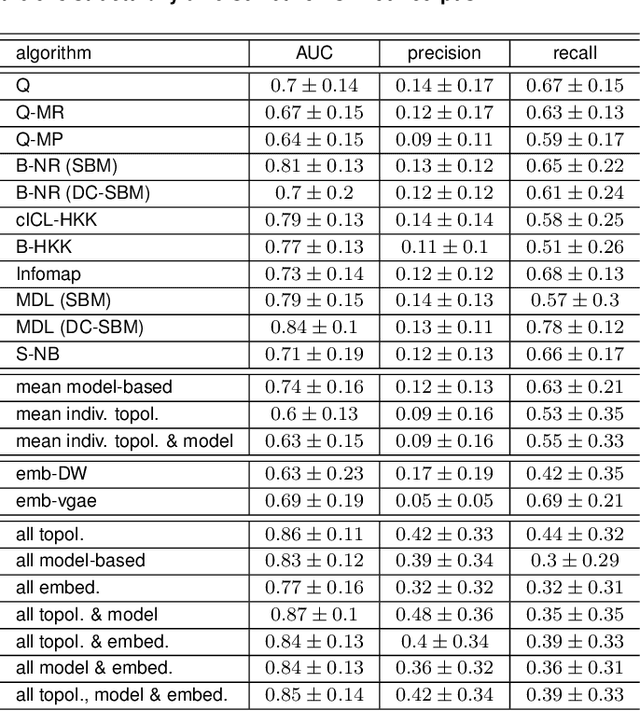
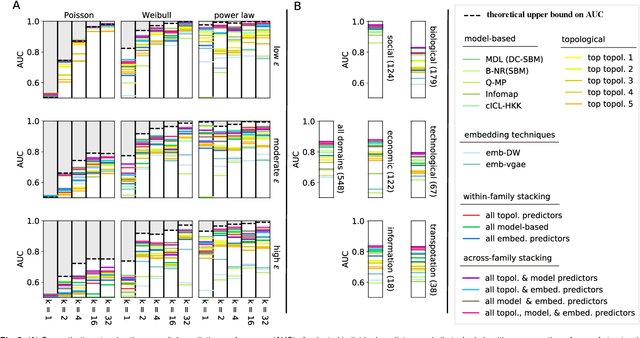
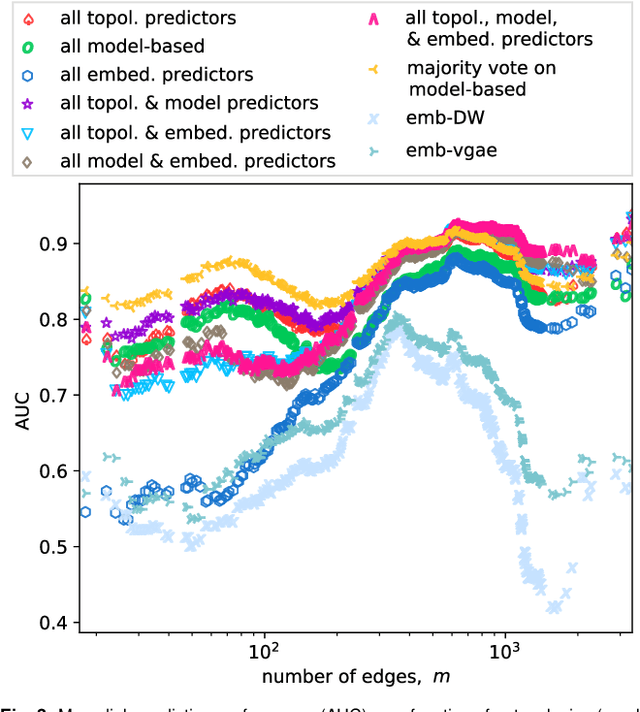
Abstract:Most real-world networks are incompletely observed. Algorithms that can accurately predict which links are missing can dramatically speedup the collection of network data and improve the validity of network models. Many algorithms now exist for predicting missing links, given a partially observed network, but it has remained unknown whether a single best predictor exists, how link predictability varies across methods and networks from different domains, and how close to optimality current methods are. We answer these questions by systematically evaluating 203 individual link predictor algorithms, representing three popular families of methods, applied to a large corpus of 548 structurally diverse networks from six scientific domains. We first show that individual algorithms exhibit a broad diversity of prediction errors, such that no one predictor or family is best, or worst, across all realistic inputs. We then exploit this diversity via meta-learning to construct a series of "stacked" models that combine predictors into a single algorithm. Applied to a broad range of synthetic networks, for which we may analytically calculate optimal performance, these stacked models achieve optimal or nearly optimal levels of accuracy. Applied to real-world networks, stacked models are also superior, but their accuracy varies strongly by domain, suggesting that link prediction may be fundamentally easier in social networks than in biological or technological networks. These results indicate that the state-of-the-art for link prediction comes from combining individual algorithms, which achieves nearly optimal predictions. We close with a brief discussion of limitations and opportunities for further improvement of these results.
Tensor Embedding: A Supervised Framework for Human Behavioral Data Mining and Prediction
Aug 31, 2018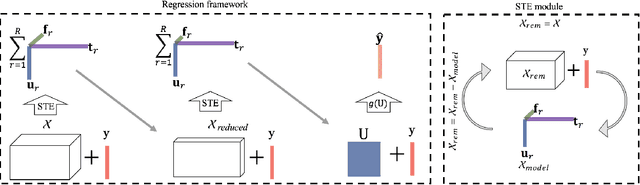
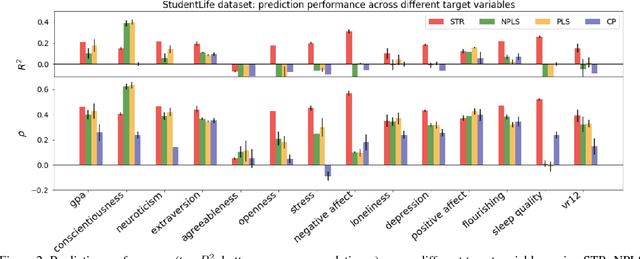
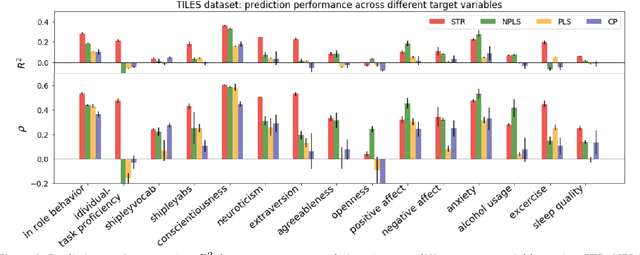
Abstract:Today's densely instrumented world offers tremendous opportunities for continuous acquisition and analysis of multimodal sensor data providing temporal characterization of an individual's behaviors. Is it possible to efficiently couple such rich sensor data with predictive modeling techniques to provide contextual, and insightful assessments of individual performance and wellbeing? Prediction of different aspects of human behavior from these noisy, incomplete, and heterogeneous bio-behavioral temporal data is a challenging problem, beyond unsupervised discovery of latent structures. We propose a Supervised Tensor Embedding (STE) algorithm for high dimension multimodal data with join decomposition of input and target variable. Furthermore, we show that features selection will help to reduce the contamination in the prediction and increase the performance. The efficiently of the methods was tested via two different real world datasets.
Evaluating Overfit and Underfit in Models of Network Community Structure
Mar 01, 2018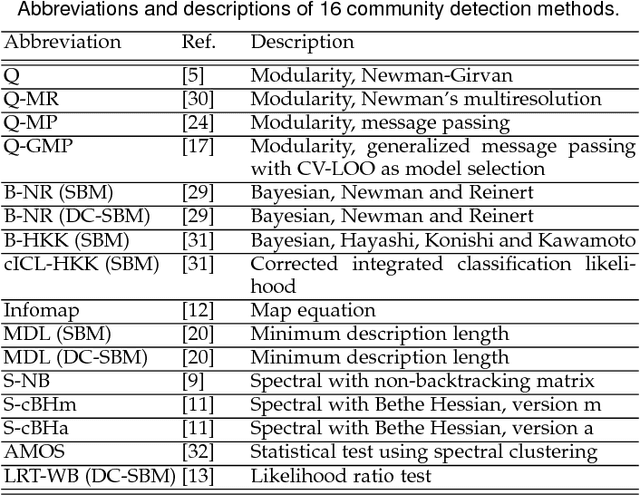
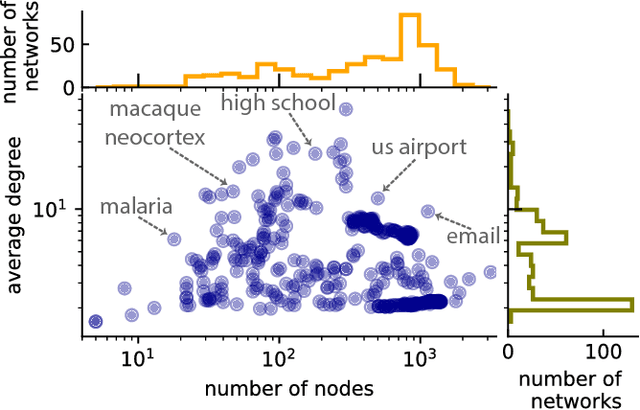
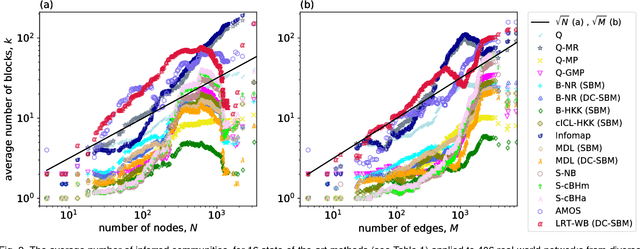
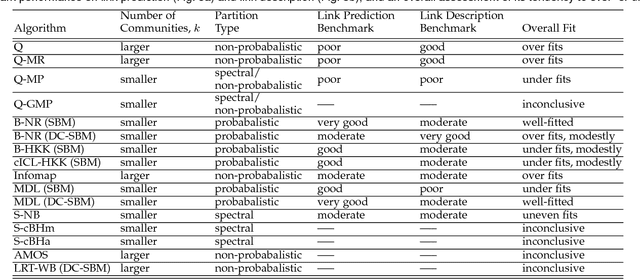
Abstract:A common data mining task on networks is community detection, which seeks an unsupervised decomposition of a network into structural groups based on statistical regularities in the network's connectivity. Although many methods exist, the No Free Lunch theorem for community detection implies that each makes some kind of tradeoff, and no algorithm can be optimal on all inputs. Thus, different algorithms will over or underfit on different inputs, finding more, fewer, or just different communities than is optimal, and evaluation methods that use a metadata partition as a ground truth will produce misleading conclusions about general accuracy. Here, we present a broad evaluation of over and underfitting in community detection, comparing the behavior of 16 state-of-the-art community detection algorithms on a novel and structurally diverse corpus of 406 real-world networks. We find that (i) algorithms vary widely both in the number of communities they find and in their corresponding composition, given the same input, (ii) algorithms can be clustered into distinct high-level groups based on similarities of their outputs on real-world networks, and (iii) these differences induce wide variation in accuracy on link prediction and link description tasks. We introduce a new diagnostic for evaluating overfitting and underfitting in practice, and use it to roughly divide community detection methods into general and specialized learning algorithms. Across methods and inputs, Bayesian techniques based on the stochastic block model and a minimum description length approach to regularization represent the best general learning approach, but can be outperformed under specific circumstances. These results introduce both a theoretically principled approach to evaluate over and underfitting in models of network community structure and a realistic benchmark by which new methods may be evaluated and compared.
Detectability thresholds and optimal algorithms for community structure in dynamic networks
Jun 19, 2015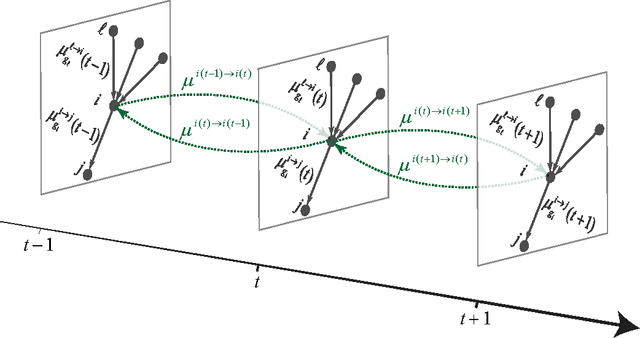
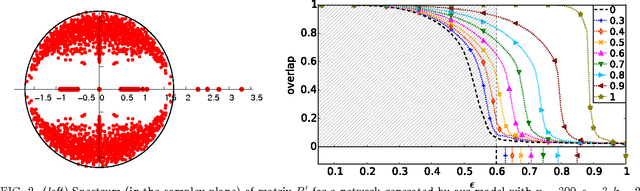
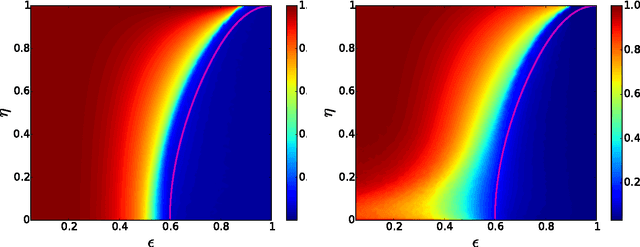
Abstract:We study the fundamental limits on learning latent community structure in dynamic networks. Specifically, we study dynamic stochastic block models where nodes change their community membership over time, but where edges are generated independently at each time step. In this setting (which is a special case of several existing models), we are able to derive the detectability threshold exactly, as a function of the rate of change and the strength of the communities. Below this threshold, we claim that no algorithm can identify the communities better than chance. We then give two algorithms that are optimal in the sense that they succeed all the way down to this limit. The first uses belief propagation (BP), which gives asymptotically optimal accuracy, and the second is a fast spectral clustering algorithm, based on linearizing the BP equations. We verify our analytic and algorithmic results via numerical simulation, and close with a brief discussion of extensions and open questions.
* 9 pages, 3 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge