Alex Pothen
QAOA Parameter Transferability for Maximum Independent Set using Graph Attention Networks
Apr 29, 2025Abstract:The quantum approximate optimization algorithm (QAOA) is one of the promising variational approaches of quantum computing to solve combinatorial optimization problems. In QAOA, variational parameters need to be optimized by solving a series of nonlinear, nonconvex optimization programs. In this work, we propose a QAOA parameter transfer scheme using Graph Attention Networks (GAT) to solve Maximum Independent Set (MIS) problems. We prepare optimized parameters for graphs of 12 and 14 vertices and use GATs to transfer their parameters to larger graphs. Additionally, we design a hybrid distributed resource-aware algorithm for MIS (HyDRA-MIS), which decomposes large problems into smaller ones that can fit onto noisy intermediate-scale quantum (NISQ) computers. We integrate our GAT-based parameter transfer approach to HyDRA-MIS and demonstrate competitive results compared to KaMIS, a state-of-the-art classical MIS solver, on graphs with several thousands vertices.
AGS-GNN: Attribute-guided Sampling for Graph Neural Networks
May 24, 2024Abstract:We propose AGS-GNN, a novel attribute-guided sampling algorithm for Graph Neural Networks (GNNs) that exploits node features and connectivity structure of a graph while simultaneously adapting for both homophily and heterophily in graphs. (In homophilic graphs vertices of the same class are more likely to be connected, and vertices of different classes tend to be linked in heterophilic graphs.) While GNNs have been successfully applied to homophilic graphs, their application to heterophilic graphs remains challenging. The best-performing GNNs for heterophilic graphs do not fit the sampling paradigm, suffer high computational costs, and are not inductive. We employ samplers based on feature-similarity and feature-diversity to select subsets of neighbors for a node, and adaptively capture information from homophilic and heterophilic neighborhoods using dual channels. Currently, AGS-GNN is the only algorithm that we know of that explicitly controls homophily in the sampled subgraph through similar and diverse neighborhood samples. For diverse neighborhood sampling, we employ submodularity, which was not used in this context prior to our work. The sampling distribution is pre-computed and highly parallel, achieving the desired scalability. Using an extensive dataset consisting of 35 small ($\le$ 100K nodes) and large (>100K nodes) homophilic and heterophilic graphs, we demonstrate the superiority of AGS-GNN compare to the current approaches in the literature. AGS-GNN achieves comparable test accuracy to the best-performing heterophilic GNNs, even outperforming methods using the entire graph for node classification. AGS-GNN also converges faster compared to methods that sample neighborhoods randomly, and can be incorporated into existing GNN models that employ node or graph sampling.
GreedyML: A Parallel Algorithm for Maximizing Submodular Functions
Mar 15, 2024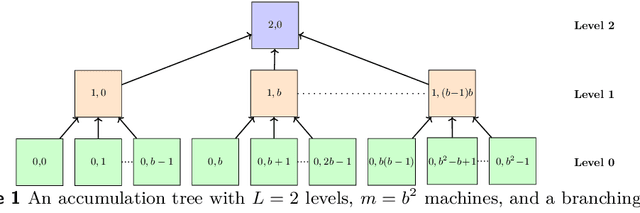
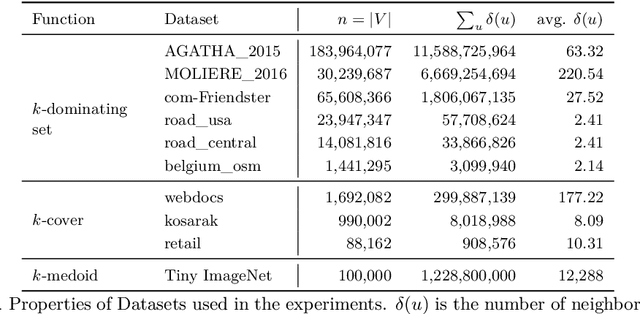
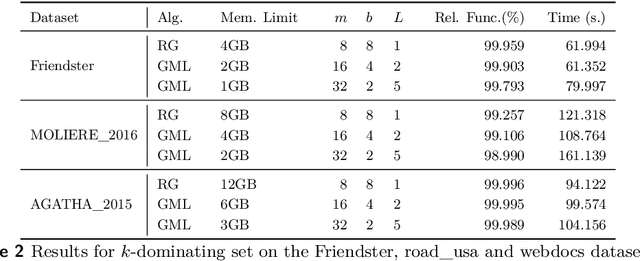
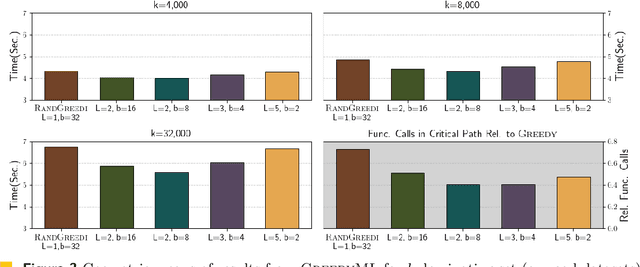
Abstract:We describe a parallel approximation algorithm for maximizing monotone submodular functions subject to hereditary constraints on distributed memory multiprocessors. Our work is motivated by the need to solve submodular optimization problems on massive data sets, for practical applications in areas such as data summarization, machine learning, and graph sparsification. Our work builds on the randomized distributed RandGreedI algorithm, proposed by Barbosa, Ene, Nguyen, and Ward (2015). This algorithm computes a distributed solution by randomly partitioning the data among all the processors and then employing a single accumulation step in which all processors send their partial solutions to one processor. However, for large problems, the accumulation step could exceed the memory available on a processor, and the processor which performs the accumulation could become a computational bottleneck. Here, we propose a generalization of the RandGreedI algorithm that employs multiple accumulation steps to reduce the memory required. We analyze the approximation ratio and the time complexity of the algorithm (in the BSP model). We also evaluate the new GreedyML algorithm on three classes of problems, and report results from massive data sets with millions of elements. The results show that the GreedyML algorithm can solve problems where the sequential Greedy and distributed RandGreedI algorithms fail due to memory constraints. For certain computationally intensive problems, the GreedyML algorithm can be faster than the RandGreedI algorithm. The observed approximation quality of the solutions computed by the GreedyML algorithm closely matches those obtained by the RandGreedI algorithm on these problems.
Generative modeling of the enteric nervous system employing point pattern analysis and graph construction
Oct 26, 2022Abstract:We describe a generative network model of the architecture of the enteric nervous system (ENS) in the colon employing data from images of human and mouse tissue samples obtained through confocal microscopy. Our models combine spatial point pattern analysis with graph generation to characterize the spatial and topological properties of the ganglia (clusters of neurons and glial cells), the inter-ganglionic connections, and the neuronal organization within the ganglia. We employ a hybrid hardcore-Strauss process for spatial patterns and a planar random graph generation for constructing the spatially embedded network. We show that our generative model may be helpful in both basic and translational studies, and it is sufficiently expressive to model the ENS architecture of individuals who vary in age and health status. Increased understanding of the ENS connectome will enable the use of neuromodulation strategies in treatment and clarify anatomic diagnostic criteria for people with bowel motility disorders.
A novel statistical methodology for quantifying the spatial arrangements of axons in peripheral nerves
Oct 18, 2022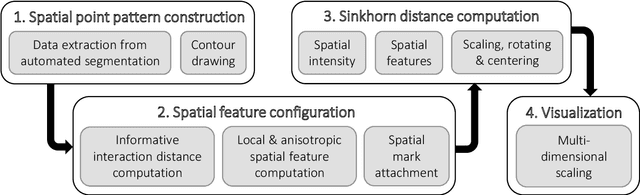
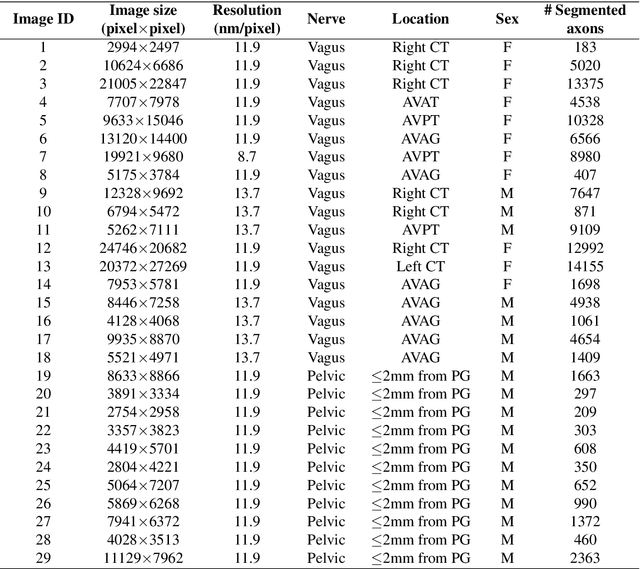
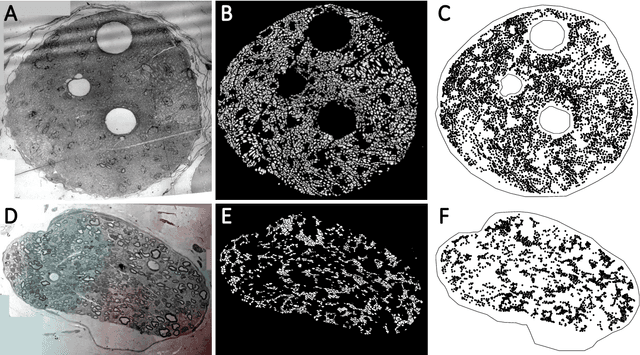
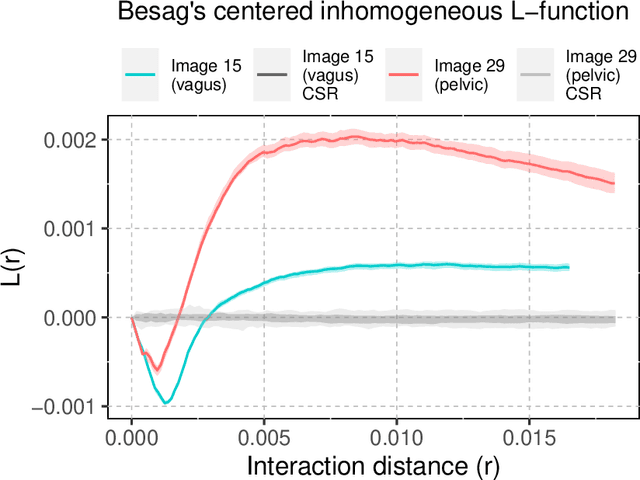
Abstract:A thorough understanding of the neuroanatomy of peripheral nerves is required for a better insight into their function and the development of neuromodulation tools and strategies. In biophysical modeling, it is commonly assumed that the complex spatial arrangement of myelinated and unmyelinated axons in peripheral nerves is random, however, in reality the axonal organization is inhomogeneous and anisotropic. Present quantitative neuroanatomy methods analyze peripheral nerves in terms of the number of axons and the morphometric characteristics of the axons, such as area and diameter. In this study, we employed spatial statistics and point process models to describe the spatial arrangement of axons and Sinkhorn distances to compute the similarities between these arrangements (in terms of first- and second-order statistics) in various vagus and pelvic nerve cross-sections. We utilized high-resolution TEM images that have been segmented using a custom-built high-throughput deep learning system based on a highly modified U-Net architecture. Our findings show a novel and innovative approach to quantifying similarities between spatial point patterns using metrics derived from the solution to the optimal transport problem. We also present a generalizable pipeline for quantitative analysis of peripheral nerve architecture. Our data demonstrate differences between male- and female-originating samples and similarities between the pelvic and abdominal vagus nerves.
V2W-BERT: A Framework for Effective Hierarchical Multiclass Classification of Software Vulnerabilities
Feb 23, 2021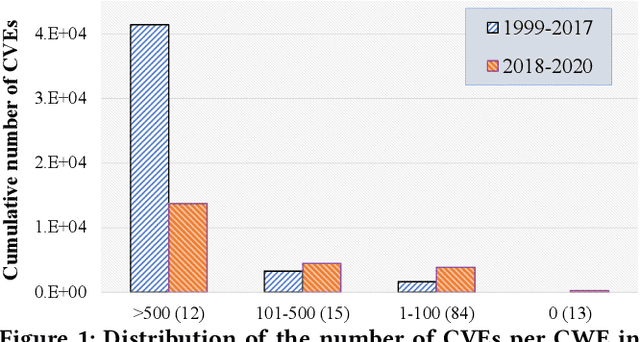
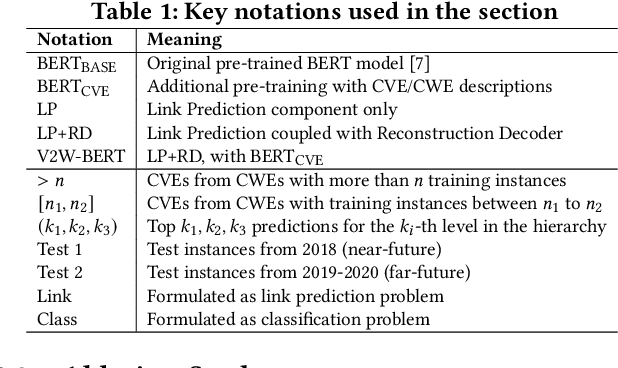
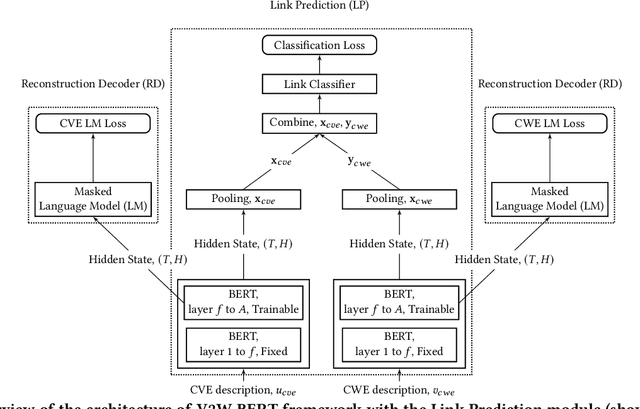
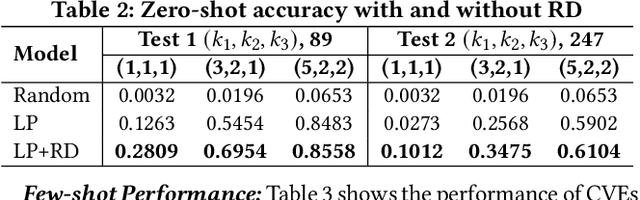
Abstract:Weaknesses in computer systems such as faults, bugs and errors in the architecture, design or implementation of software provide vulnerabilities that can be exploited by attackers to compromise the security of a system. Common Weakness Enumerations (CWE) are a hierarchically designed dictionary of software weaknesses that provide a means to understand software flaws, potential impact of their exploitation, and means to mitigate these flaws. Common Vulnerabilities and Exposures (CVE) are brief low-level descriptions that uniquely identify vulnerabilities in a specific product or protocol. Classifying or mapping of CVEs to CWEs provides a means to understand the impact and mitigate the vulnerabilities. Since manual mapping of CVEs is not a viable option, automated approaches are desirable but challenging. We present a novel Transformer-based learning framework (V2W-BERT) in this paper. By using ideas from natural language processing, link prediction and transfer learning, our method outperforms previous approaches not only for CWE instances with abundant data to train, but also rare CWE classes with little or no data to train. Our approach also shows significant improvements in using historical data to predict links for future instances of CVEs, and therefore, provides a viable approach for practical applications. Using data from MITRE and National Vulnerability Database, we achieve up to 97% prediction accuracy for randomly partitioned data and up to 94% prediction accuracy in temporally partitioned data. We believe that our work will influence the design of better methods and training models, as well as applications to solve increasingly harder problems in cybersecurity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge