Alessandra Lumini
Feature transforms for image data augmentation
Jan 24, 2022
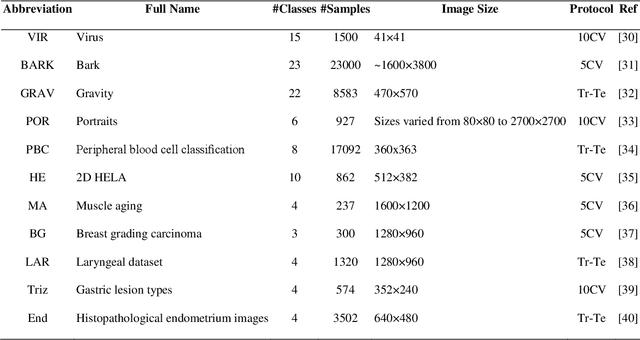
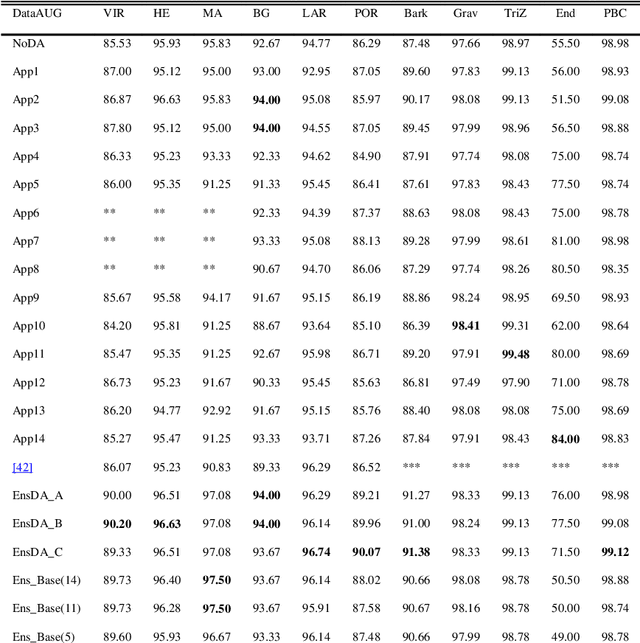
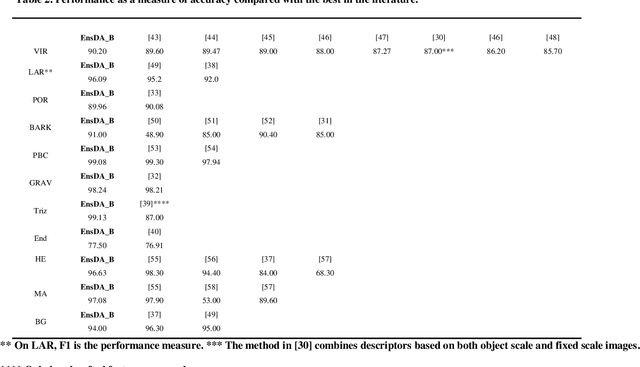
Abstract:A problem with Convolutional Neural Networks (CNNs) is that they require large datasets to obtain adequate robustness; on small datasets, they are prone to overfitting. Many methods have been proposed to overcome this shortcoming with CNNs. In cases where additional samples cannot easily be collected, a common approach is to generate more data points from existing data using an augmentation technique. In image classification, many augmentation approaches utilize simple image manipulation algorithms. In this work, we build ensembles on the data level by adding images generated by combining fourteen augmentation approaches, three of which are proposed here for the first time. These novel methods are based on the Fourier Transform (FT), the Radon Transform (RT) and the Discrete Cosine Transform (DCT). Pretrained ResNet50 networks are finetuned on training sets that include images derived from each augmentation method. These networks and several fusions are evaluated and compared across eleven benchmarks. Results show that building ensembles on the data level by combining different data augmentation methods produce classifiers that not only compete competitively against the state-of-the-art but often surpass the best approaches reported in the literature.
Deep ensembles in bioimage segmentation
Dec 24, 2021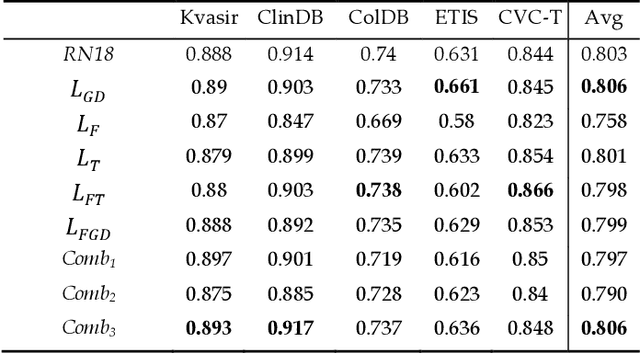
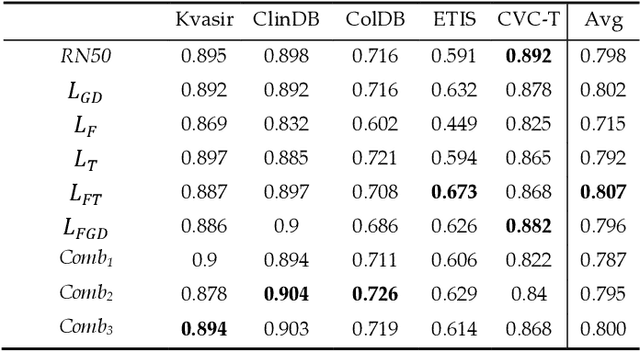
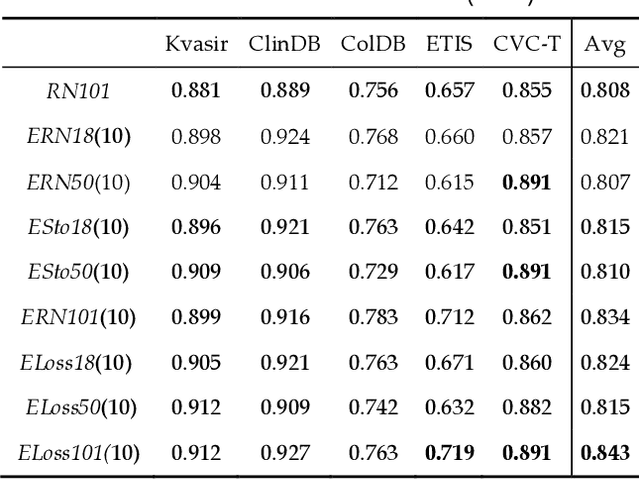
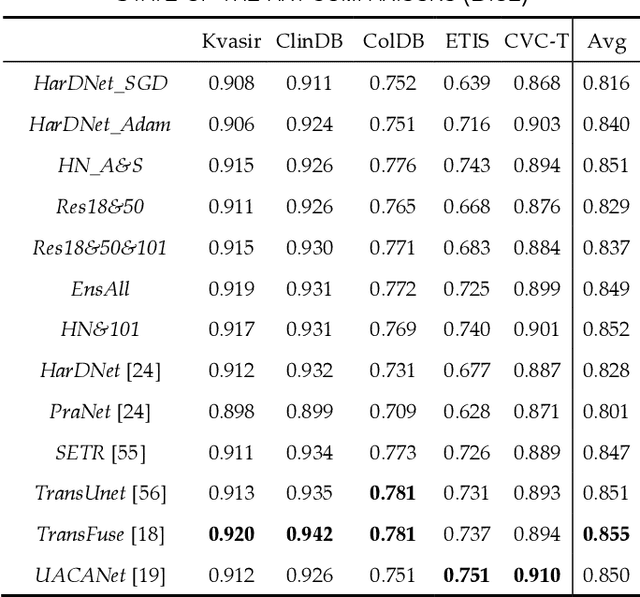
Abstract:Semantic segmentation consists in classifying each pixel of an image by assigning it to a specific label chosen from a set of all the available ones. During the last few years, a lot of attention shifted to this kind of task. Many computer vision researchers tried to apply autoencoder structures to develop models that can learn the semantics of the image as well as a low-level representation of it. In an autoencoder architecture, given an input, an encoder computes a low dimensional representation of the input that is then used by a decoder to reconstruct the original data. In this work, we propose an ensemble of convolutional neural networks (CNNs). In ensemble methods, many different models are trained and then used for classification, the ensemble aggregates the outputs of the single classifiers. The approach leverages on differences of various classifiers to improve the performance of the whole system. Diversity among the single classifiers is enforced by using different loss functions. In particular, we present a new loss function that results from the combination of Dice and Structural Similarity Index. The proposed ensemble is implemented by combining different backbone networks using the DeepLabV3+ and HarDNet environment. The proposal is evaluated through an extensive empirical evaluation on two real-world scenarios: polyp and skin segmentation. All the code is available online at https://github.com/LorisNanni.
Gated recurrent units and temporal convolutional network for multilabel classification
Oct 09, 2021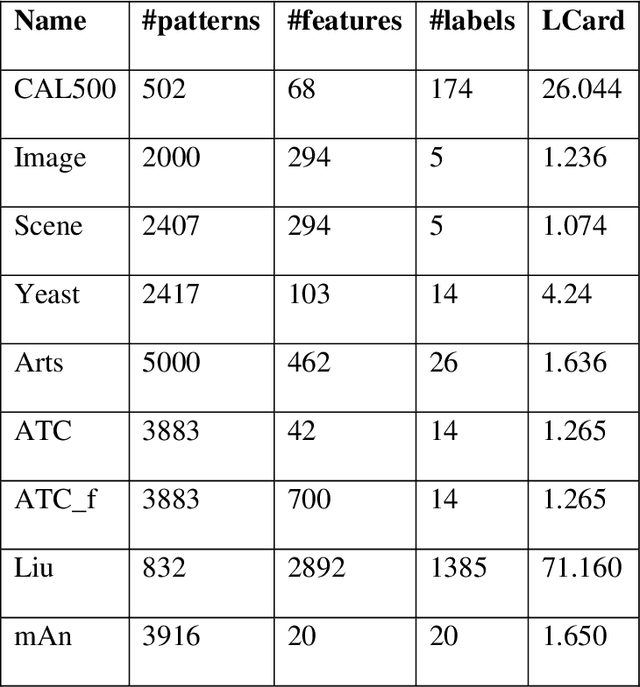
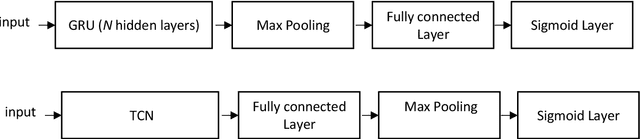
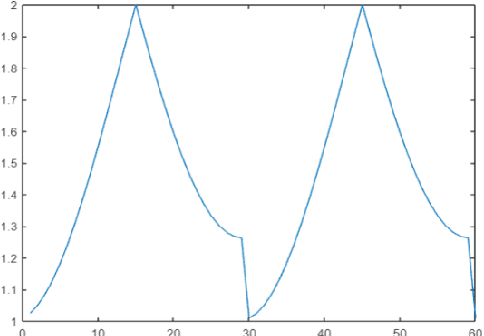
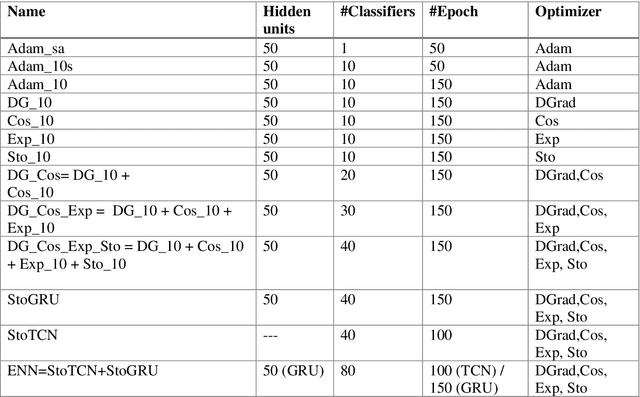
Abstract:Multilabel learning tackles the problem of associating a sample with multiple class labels. This work proposes a new ensemble method for managing multilabel classification: the core of the proposed approach combines a set of gated recurrent units and temporal convolutional neural networks trained with variants of the Adam optimization approach. Multiple Adam variants, including novel one proposed here, are compared and tested; these variants are based on the difference between present and past gradients, with step size adjusted for each parameter. The proposed neural network approach is also combined with Incorporating Multiple Clustering Centers (IMCC), which further boosts classification performance. Multiple experiments on nine data sets representing a wide variety of multilabel tasks demonstrate the robustness of our best ensemble, which is shown to outperform the state-of-the-art. The MATLAB code for generating the best ensembles in the experimental section will be available at https://github.com/LorisNanni.
High performing ensemble of convolutional neural networks for insect pest image detection
Aug 28, 2021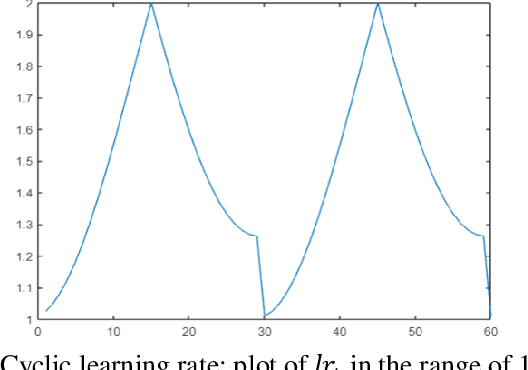
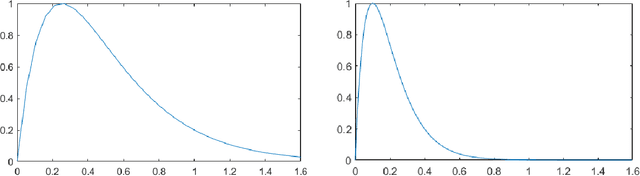
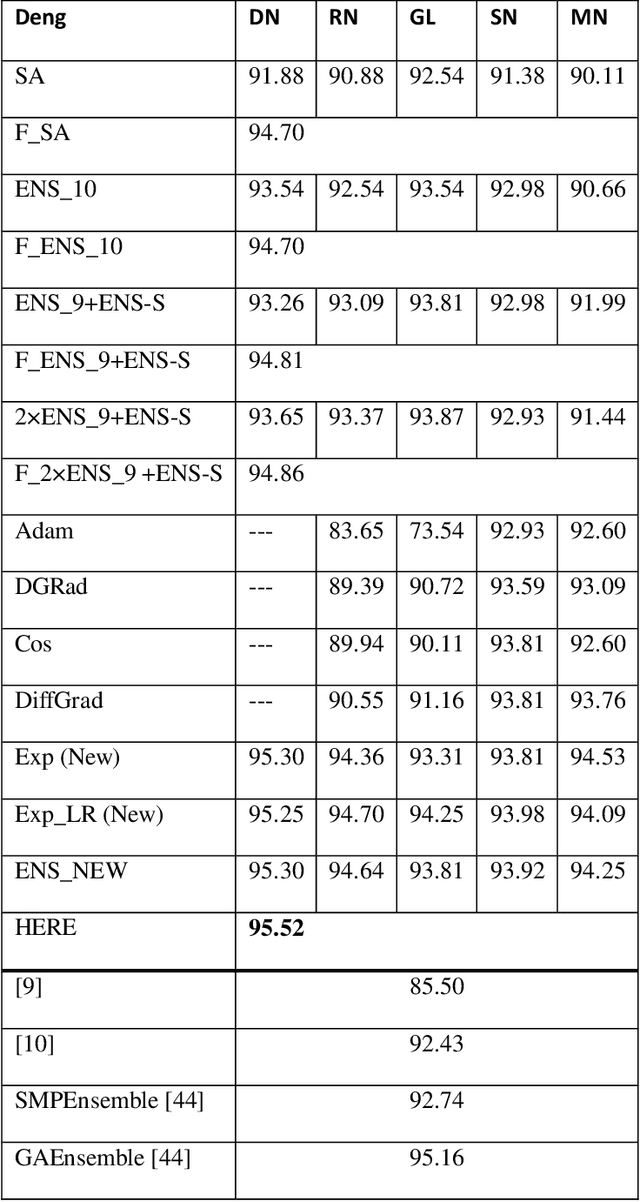

Abstract:Pest infestation is a major cause of crop damage and lost revenues worldwide. Automatic identification of invasive insects would greatly speedup the identification of pests and expedite their removal. In this paper, we generate ensembles of CNNs based on different topologies (ResNet50, GoogleNet, ShuffleNet, MobileNetv2, and DenseNet201) altered by random selection from a simple set of data augmentation methods or optimized with different Adam variants for pest identification. Two new Adam algorithms for deep network optimization based on DGrad are proposed that introduce a scaling factor in the learning rate. Sets of the five CNNs that vary in either data augmentation or the type of Adam optimization were trained on both the Deng (SMALL) and the large IP102 pest data sets. Ensembles were compared and evaluated using three performance indicators. The best performing ensemble, which combined the CNNs using the different augmentation methods and the two new Adam variants proposed here, achieved state of the art on both insect data sets: 95.52% on Deng and 73.46% on IP102, a score on Deng that competed with human expert classifications. Additional tests were performed on data sets for medical imagery classification that further validated the robustness and power of the proposed Adam optimization variants. All MATLAB source code is available at https://github.com/LorisNanni/.
Deep ensembles based on Stochastic Activation Selection for Polyp Segmentation
Apr 07, 2021
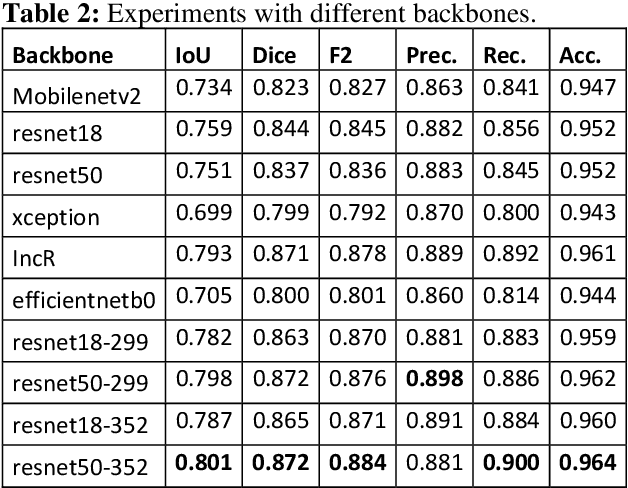
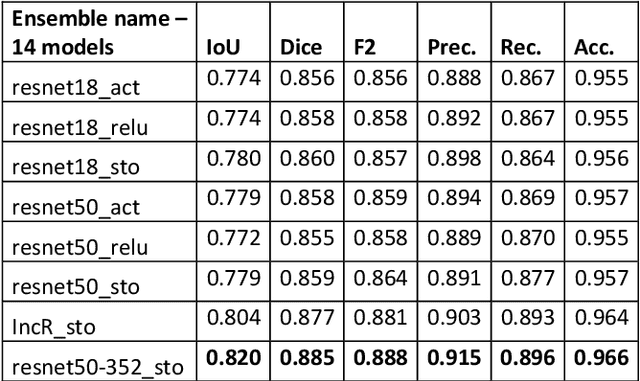
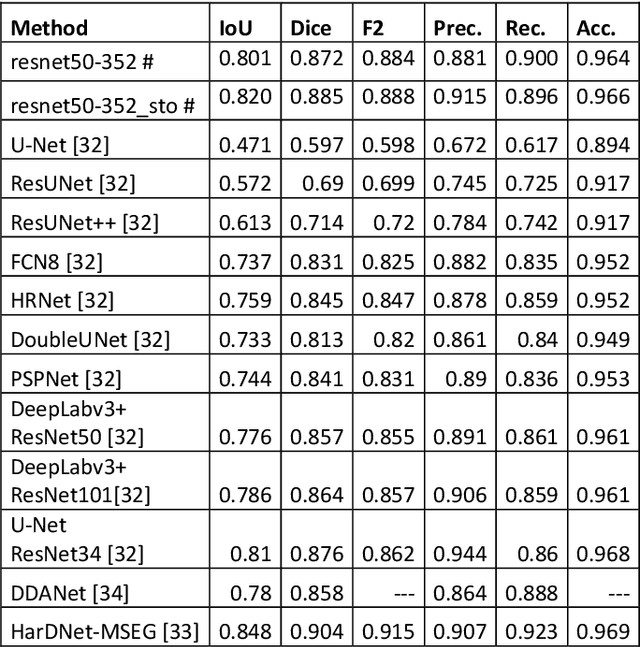
Abstract:Semantic segmentation has a wide array of applications ranging from medical-image analysis, scene understanding, autonomous driving and robotic navigation. This work deals with medical image segmentation and in particular with accurate polyp detection and segmentation during colonoscopy examinations. Several convolutional neural network architectures have been proposed to effectively deal with this task and with the problem of segmenting objects at different scale input. The basic architecture in image segmentation consists of an encoder and a decoder: the first uses convolutional filters to extract features from the image, the second is responsible for generating the final output. In this work, we compare some variant of the DeepLab architecture obtained by varying the decoder backbone. We compare several decoder architectures, including ResNet, Xception, EfficentNet, MobileNet and we perturb their layers by substituting ReLU activation layers with other functions. The resulting methods are used to create deep ensembles which are shown to be very effective. Our experimental evaluations show that our best ensemble produces good segmentation results by achieving high evaluation scores with a dice coefficient of 0.884, and a mean Intersection over Union (mIoU) of 0.818 for the Kvasir-SEG dataset. To improve reproducibility and research efficiency the MATLAB source code used for this research is available at GitHub: https://github.com/LorisNanni.
Exploiting Adam-like Optimization Algorithms to Improve the Performance of Convolutional Neural Networks
Mar 26, 2021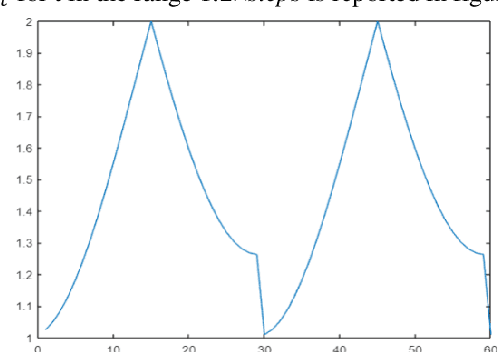
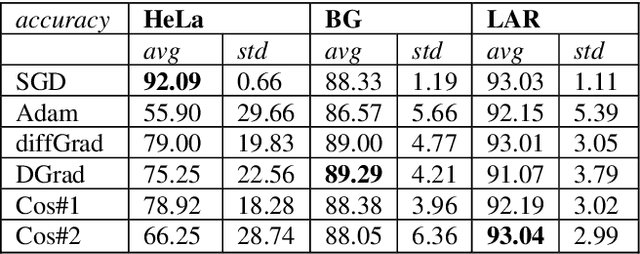
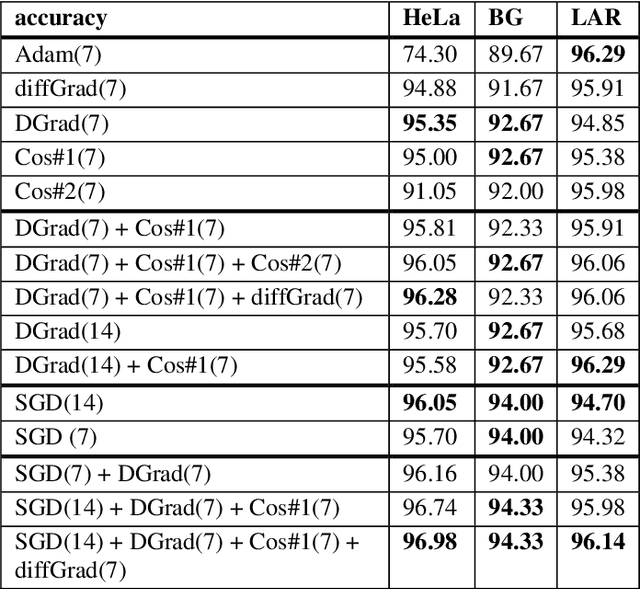
Abstract:Stochastic gradient descent (SGD) is the main approach for training deep networks: it moves towards the optimum of the cost function by iteratively updating the parameters of a model in the direction of the gradient of the loss evaluated on a minibatch. Several variants of SGD have been proposed to make adaptive step sizes for each parameter (adaptive gradient) and take into account the previous updates (momentum). Among several alternative of SGD the most popular are AdaGrad, AdaDelta, RMSProp and Adam which scale coordinates of the gradient by square roots of some form of averaging of the squared coordinates in the past gradients and automatically adjust the learning rate on a parameter basis. In this work, we compare Adam based variants based on the difference between the present and the past gradients, the step size is adjusted for each parameter. We run several tests benchmarking proposed methods using medical image data. The experiments are performed using ResNet50 architecture neural network. Moreover, we have tested ensemble of networks and the fusion with ResNet50 trained with stochastic gradient descent. To combine the set of ResNet50 the simple sum rule has been applied. Proposed ensemble obtains very high performance, it obtains accuracy comparable or better than actual state of the art. To improve reproducibility and research efficiency the MATLAB source code used for this research is available at GitHub: https://github.com/LorisNanni.
Neural networks for Anatomical Therapeutic Chemical (ATC)
Jan 22, 2021
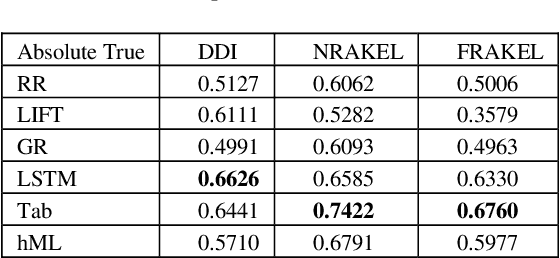
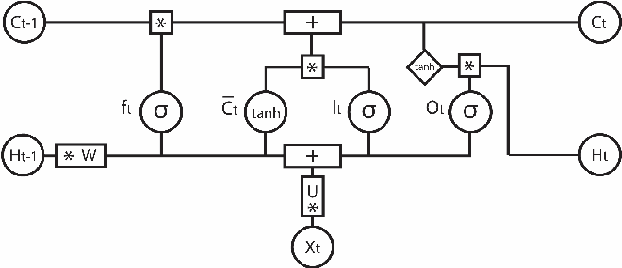
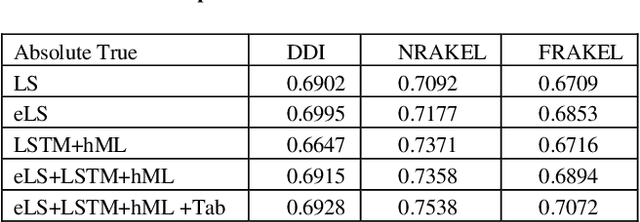
Abstract:Motivation: Automatic Anatomical Therapeutic Chemical (ATC) classification is a critical and highly competitive area of research in bioinformatics because of its potential for expediting drug develop-ment and research. Predicting an unknown compound's therapeutic and chemical characteristics ac-cording to how these characteristics affect multiple organs/systems makes automatic ATC classifica-tion a challenging multi-label problem. Results: In this work, we propose combining multiple multi-label classifiers trained on distinct sets of features, including sets extracted from a Bidirectional Long Short-Term Memory Network (BiLSTM). Experiments demonstrate the power of this approach, which is shown to outperform the best methods reported in the literature, including the state-of-the-art developed by the fast.ai research group. Availability: All source code developed for this study is available at https://github.com/LorisNanni. Contact: loris.nanni@unipd.it
Comparisons among different stochastic selection of activation layers for convolutional neural networks for healthcare
Nov 24, 2020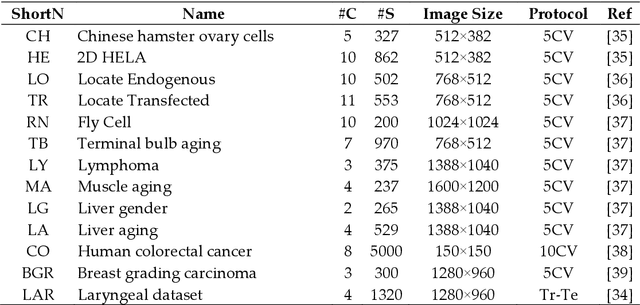

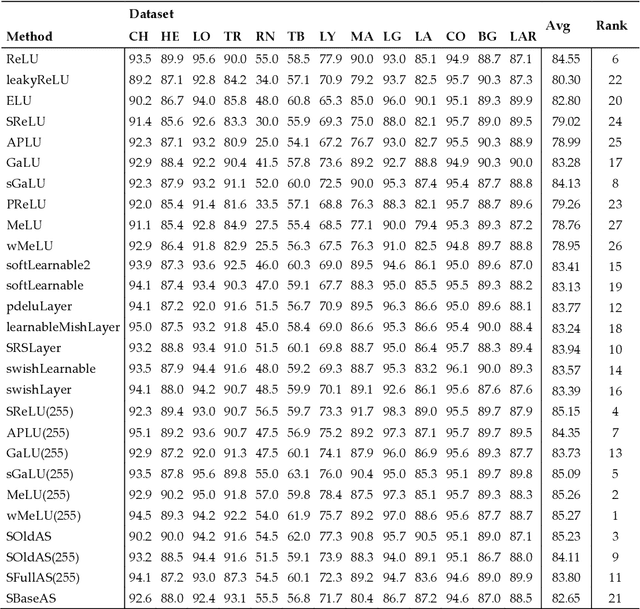
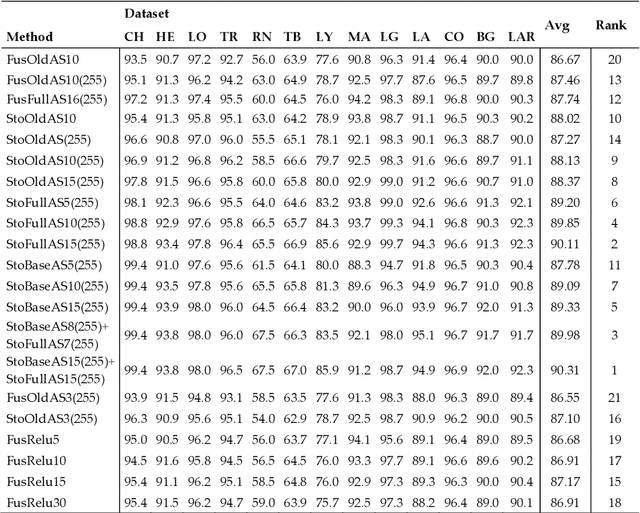
Abstract:Classification of biological images is an important task with crucial application in many fields, such as cell phenotypes recognition, detection of cell organelles and histopathological classification, and it might help in early medical diagnosis, allowing automatic disease classification without the need of a human expert. In this paper we classify biomedical images using ensembles of neural networks. We create this ensemble using a ResNet50 architecture and modifying its activation layers by substituting ReLUs with other functions. We select our activations among the following ones: ReLU, leaky ReLU, Parametric ReLU, ELU, Adaptive Piecewice Linear Unit, S-Shaped ReLU, Swish , Mish, Mexican Linear Unit, Gaussian Linear Unit, Parametric Deformable Linear Unit, Soft Root Sign (SRS) and others. As a baseline, we used an ensemble of neural networks that only use ReLU activations. We tested our networks on several small and medium sized biomedical image datasets. Our results prove that our best ensemble obtains a better performance than the ones of the naive approaches. In order to encourage the reproducibility of this work, the MATLAB code of all the experiments will be shared at https://github.com/LorisNanni.
Deep learning for Plankton and Coral Classification
Aug 15, 2019

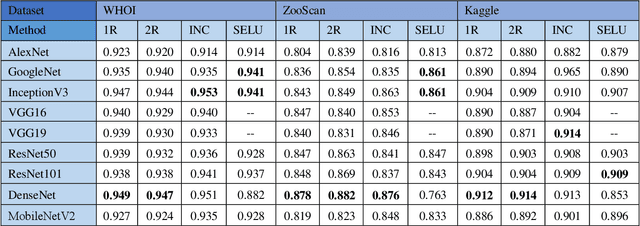
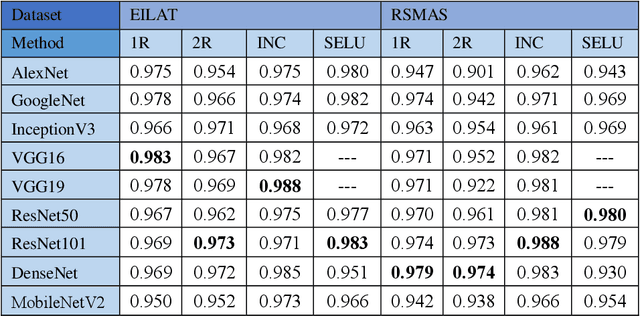
Abstract:Oceans are the essential lifeblood of the Earth: they provide over 70% of the oxygen and over 97% of the water. Plankton and corals are two of the most fundamental components of ocean ecosystems, the former due to their function at many levels of the oceans food chain, the latter because they provide spawning and nursery grounds to many fish populations. Studying and monitoring plankton distribution and coral reefs is vital for environment protection. In the last years there has been a massive proliferation of digital imagery for the monitoring of underwater ecosystems and much research is concentrated on the automated recognition of plankton and corals. In this paper, we present a study about an automated system for monitoring of underwater ecosystems. The system here proposed is based on the fusion of different deep learning methods. We study how to create an ensemble based of different CNN models, fine tuned on several datasets with the aim of exploiting their diversity. The aim of our study is to experiment the possibility of fine-tuning pretrained CNN for underwater imagery analysis, the opportunity of using different datasets for pretraining models, the possibility to design an ensemble using the same architecture with small variations in the training procedure. The experimental results are very encouraging, our experiments performed on 5 well-knowns datasets (3 plankton and 2 coral datasets) show that the fusion of such different CNN models in a heterogeneous ensemble grants a substantial performance improvement with respect to other state-of-the-art approaches in all the tested problems. One of the main contributions of this work is a wide experimental evaluation of famous CNN architectures to report performance of both single CNN and ensemble of CNNs in different problems. Moreover, we show how to create an ensemble which improves the performance of the best single model.
Learning morphological operators for skin detection
Aug 09, 2019

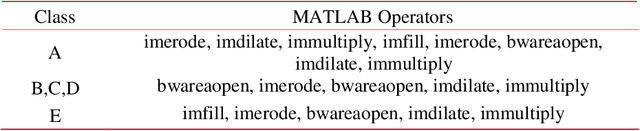
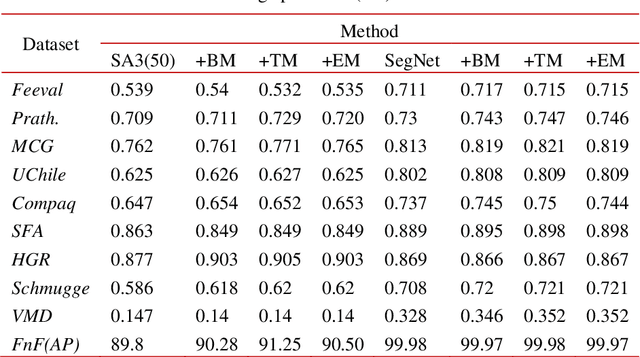
Abstract:In this work we propose a novel post processing approach for skin detectors based on trained morphological operators. The first step, consisting in skin segmentation is performed according to an existing skin detection approach is performed for skin segmentation, then a second step is carried out consisting in the application of a set of morphological operators to refine the resulting mask. Extensive experimental evaluation performed considering two different detection approaches (one based on deep learning and a handcrafted one) carried on 10 different datasets confirms the quality of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge