Sheryl Brahnam
Feature transforms for image data augmentation
Jan 24, 2022
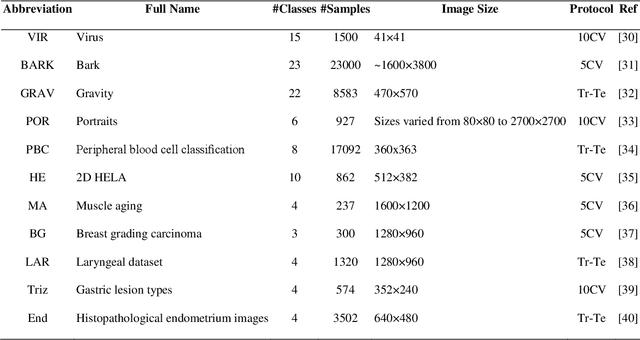
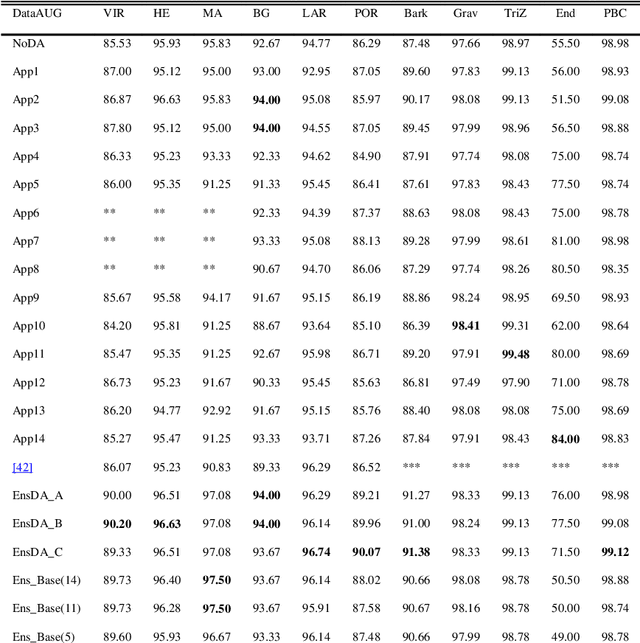
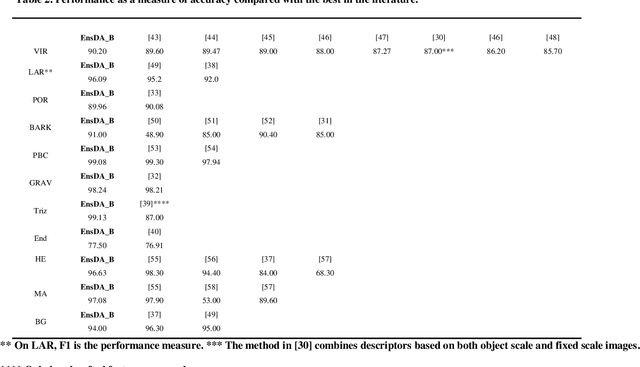
Abstract:A problem with Convolutional Neural Networks (CNNs) is that they require large datasets to obtain adequate robustness; on small datasets, they are prone to overfitting. Many methods have been proposed to overcome this shortcoming with CNNs. In cases where additional samples cannot easily be collected, a common approach is to generate more data points from existing data using an augmentation technique. In image classification, many augmentation approaches utilize simple image manipulation algorithms. In this work, we build ensembles on the data level by adding images generated by combining fourteen augmentation approaches, three of which are proposed here for the first time. These novel methods are based on the Fourier Transform (FT), the Radon Transform (RT) and the Discrete Cosine Transform (DCT). Pretrained ResNet50 networks are finetuned on training sets that include images derived from each augmentation method. These networks and several fusions are evaluated and compared across eleven benchmarks. Results show that building ensembles on the data level by combining different data augmentation methods produce classifiers that not only compete competitively against the state-of-the-art but often surpass the best approaches reported in the literature.
Deep ensembles in bioimage segmentation
Dec 24, 2021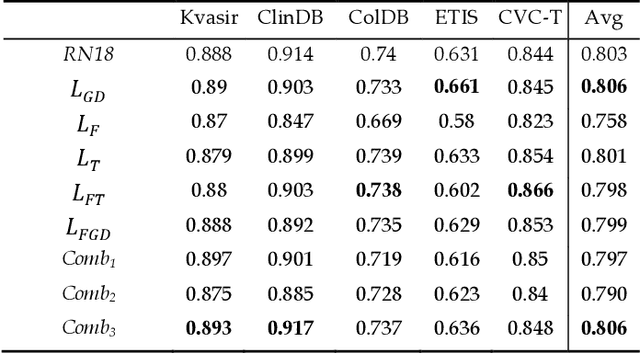
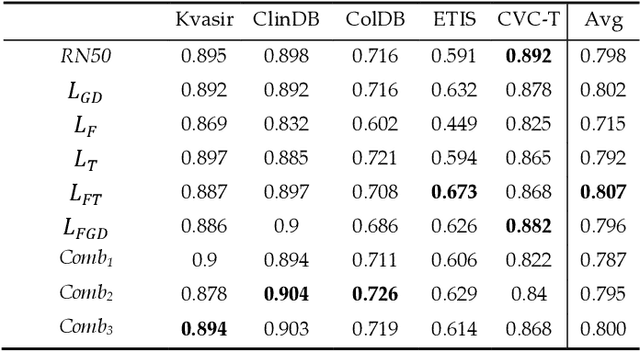
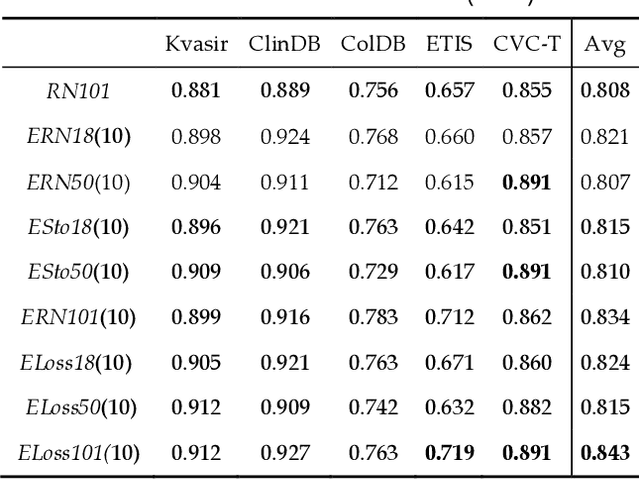
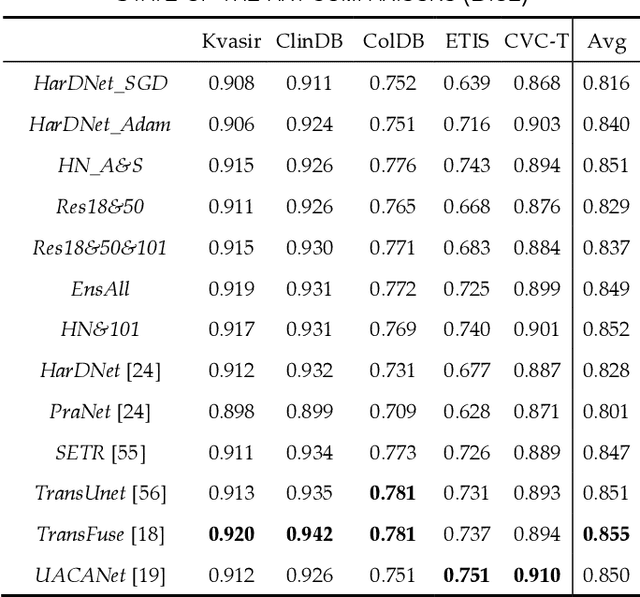
Abstract:Semantic segmentation consists in classifying each pixel of an image by assigning it to a specific label chosen from a set of all the available ones. During the last few years, a lot of attention shifted to this kind of task. Many computer vision researchers tried to apply autoencoder structures to develop models that can learn the semantics of the image as well as a low-level representation of it. In an autoencoder architecture, given an input, an encoder computes a low dimensional representation of the input that is then used by a decoder to reconstruct the original data. In this work, we propose an ensemble of convolutional neural networks (CNNs). In ensemble methods, many different models are trained and then used for classification, the ensemble aggregates the outputs of the single classifiers. The approach leverages on differences of various classifiers to improve the performance of the whole system. Diversity among the single classifiers is enforced by using different loss functions. In particular, we present a new loss function that results from the combination of Dice and Structural Similarity Index. The proposed ensemble is implemented by combining different backbone networks using the DeepLabV3+ and HarDNet environment. The proposal is evaluated through an extensive empirical evaluation on two real-world scenarios: polyp and skin segmentation. All the code is available online at https://github.com/LorisNanni.
Gated recurrent units and temporal convolutional network for multilabel classification
Oct 09, 2021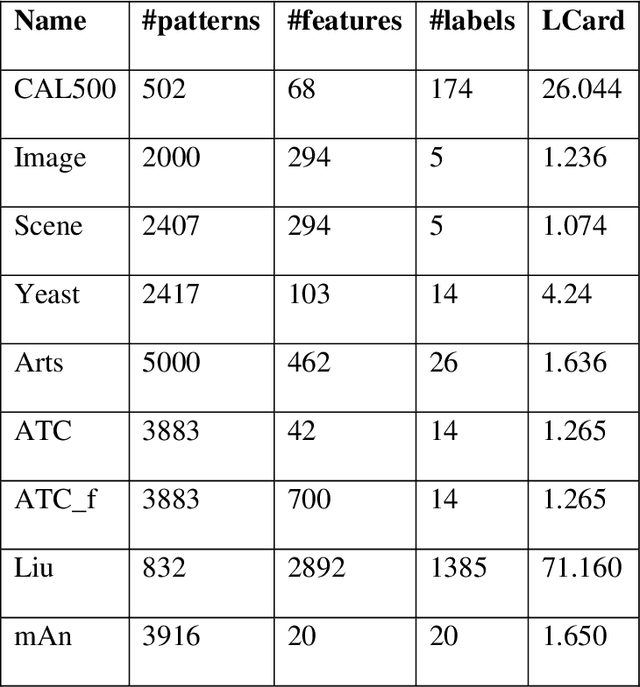
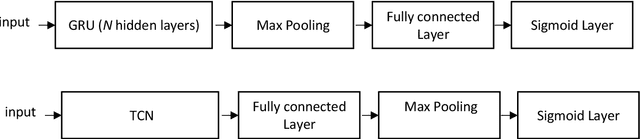
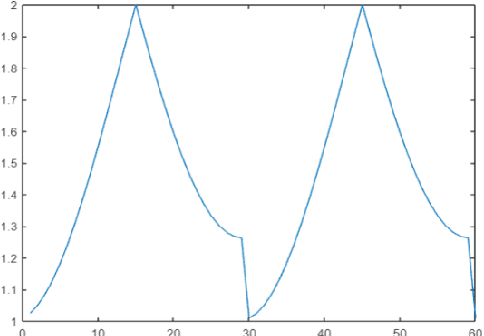
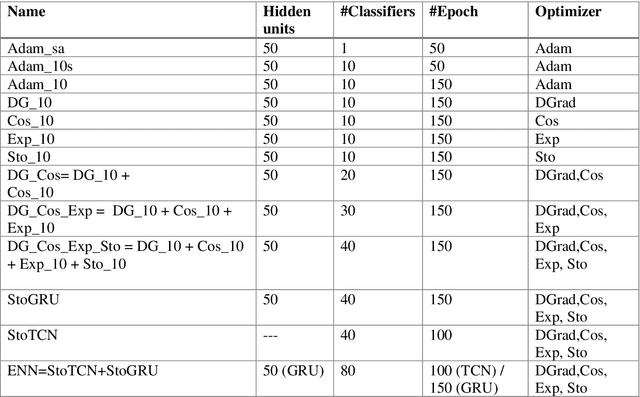
Abstract:Multilabel learning tackles the problem of associating a sample with multiple class labels. This work proposes a new ensemble method for managing multilabel classification: the core of the proposed approach combines a set of gated recurrent units and temporal convolutional neural networks trained with variants of the Adam optimization approach. Multiple Adam variants, including novel one proposed here, are compared and tested; these variants are based on the difference between present and past gradients, with step size adjusted for each parameter. The proposed neural network approach is also combined with Incorporating Multiple Clustering Centers (IMCC), which further boosts classification performance. Multiple experiments on nine data sets representing a wide variety of multilabel tasks demonstrate the robustness of our best ensemble, which is shown to outperform the state-of-the-art. The MATLAB code for generating the best ensembles in the experimental section will be available at https://github.com/LorisNanni.
High performing ensemble of convolutional neural networks for insect pest image detection
Aug 28, 2021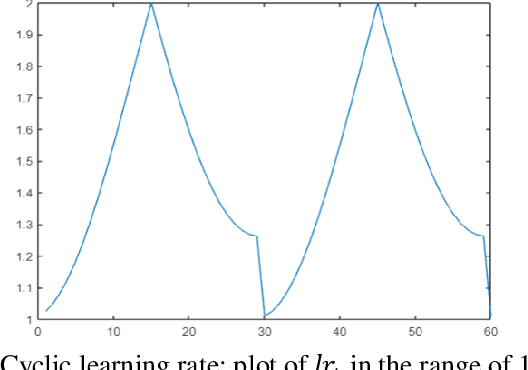
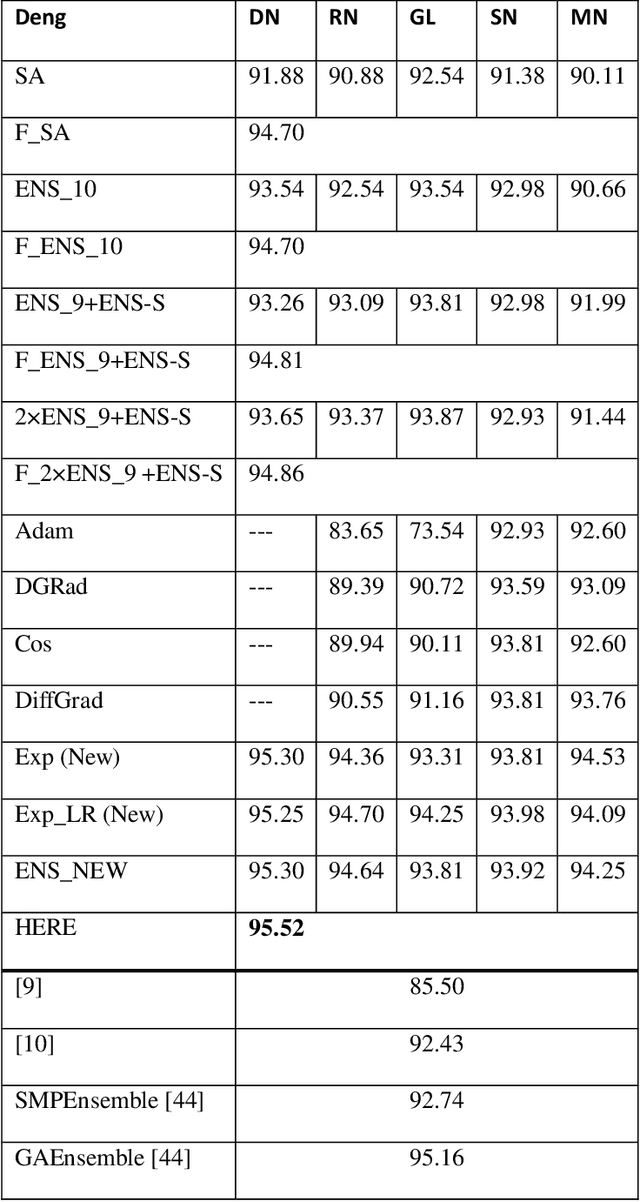

Abstract:Pest infestation is a major cause of crop damage and lost revenues worldwide. Automatic identification of invasive insects would greatly speedup the identification of pests and expedite their removal. In this paper, we generate ensembles of CNNs based on different topologies (ResNet50, GoogleNet, ShuffleNet, MobileNetv2, and DenseNet201) altered by random selection from a simple set of data augmentation methods or optimized with different Adam variants for pest identification. Two new Adam algorithms for deep network optimization based on DGrad are proposed that introduce a scaling factor in the learning rate. Sets of the five CNNs that vary in either data augmentation or the type of Adam optimization were trained on both the Deng (SMALL) and the large IP102 pest data sets. Ensembles were compared and evaluated using three performance indicators. The best performing ensemble, which combined the CNNs using the different augmentation methods and the two new Adam variants proposed here, achieved state of the art on both insect data sets: 95.52% on Deng and 73.46% on IP102, a score on Deng that competed with human expert classifications. Additional tests were performed on data sets for medical imagery classification that further validated the robustness and power of the proposed Adam optimization variants. All MATLAB source code is available at https://github.com/LorisNanni/.
Deep Features for training Support Vector Machine
Apr 08, 2021
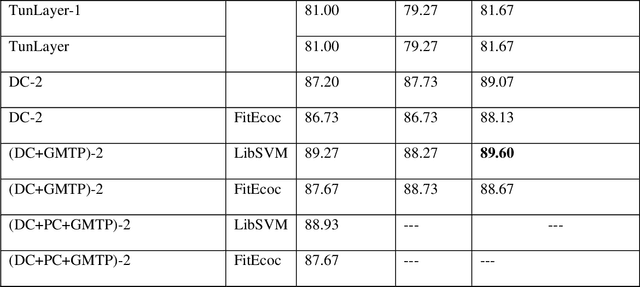

Abstract:Features play a crucial role in computer vision. Initially designed to detect salient elements by means of handcrafted algorithms, features are now often learned by different layers in Convolutional Neural Networks (CNNs). This paper develops a generic computer vision system based on features extracted from trained CNNs. Multiple learned features are combined into a single structure to work on different image classification tasks. The proposed system was experimentally derived by testing several approaches for extracting features from the inner layers of CNNs and using them as inputs to SVMs that are then combined by sum rule. Dimensionality reduction techniques are used to reduce the high dimensionality of inner layers. The resulting vision system is shown to significantly boost the performance of standard CNNs across a large and diverse collection of image data sets. An ensemble of different topologies using the same approach obtains state-of-the-art results on a virus data set.
Comparison of different convolutional neural network activation functions and methods for building ensembles
Apr 02, 2021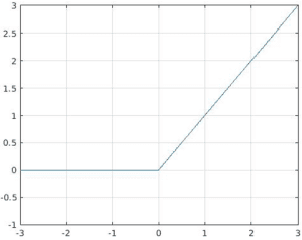
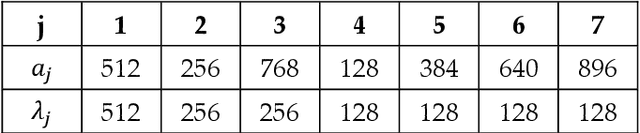
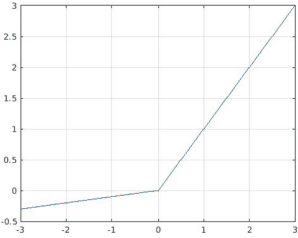
Abstract:Recently, much attention has been devoted to finding highly efficient and powerful activation functions for CNN layers. Because activation functions inject different nonlinearities between layers that affect performance, varying them is one method for building robust ensembles of CNNs. The objective of this study is to examine the performance of CNN ensembles made with different activation functions, including six new ones presented here: 2D Mexican ReLU, TanELU, MeLU+GaLU, Symmetric MeLU, Symmetric GaLU, and Flexible MeLU. The highest performing ensemble was built with CNNs having different activation layers that randomly replaced the standard ReLU. A comprehensive evaluation of the proposed approach was conducted across fifteen biomedical data sets representing various classification tasks. The proposed method was tested on two basic CNN architectures: Vgg16 and ResNet50. Results demonstrate the superiority in performance of this approach. The MATLAB source code for this study will be available at https://github.com/LorisNanni.
Neural networks for Anatomical Therapeutic Chemical (ATC)
Jan 22, 2021
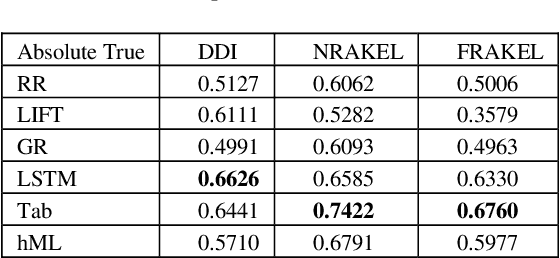
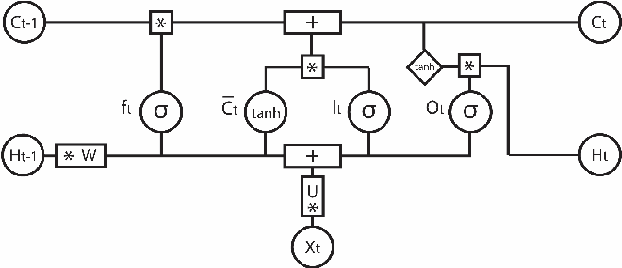
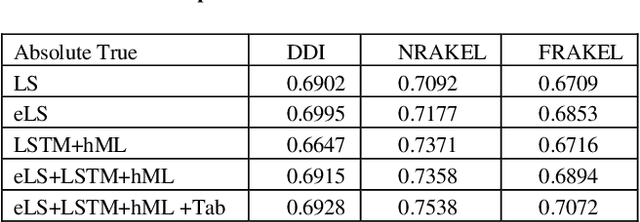
Abstract:Motivation: Automatic Anatomical Therapeutic Chemical (ATC) classification is a critical and highly competitive area of research in bioinformatics because of its potential for expediting drug develop-ment and research. Predicting an unknown compound's therapeutic and chemical characteristics ac-cording to how these characteristics affect multiple organs/systems makes automatic ATC classifica-tion a challenging multi-label problem. Results: In this work, we propose combining multiple multi-label classifiers trained on distinct sets of features, including sets extracted from a Bidirectional Long Short-Term Memory Network (BiLSTM). Experiments demonstrate the power of this approach, which is shown to outperform the best methods reported in the literature, including the state-of-the-art developed by the fast.ai research group. Availability: All source code developed for this study is available at https://github.com/LorisNanni. Contact: loris.nanni@unipd.it
An Ensemble of Convolutional Neural Networks for Audio Classification
Jul 15, 2020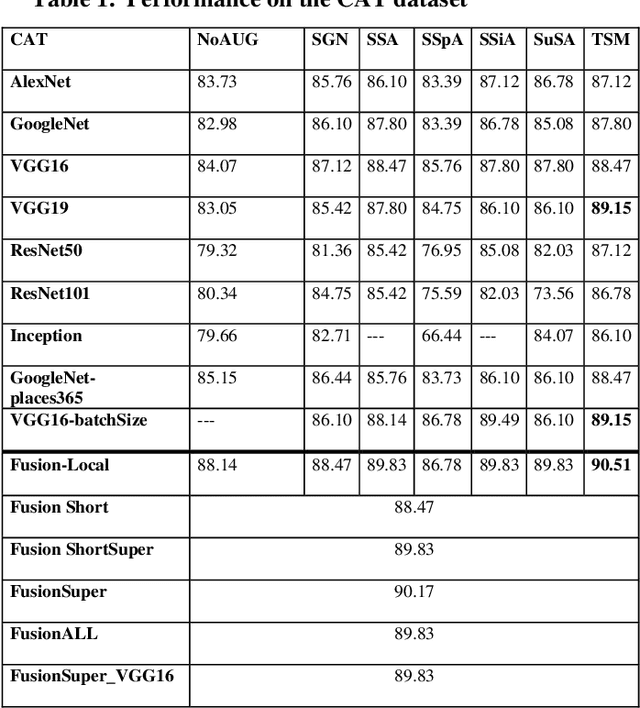

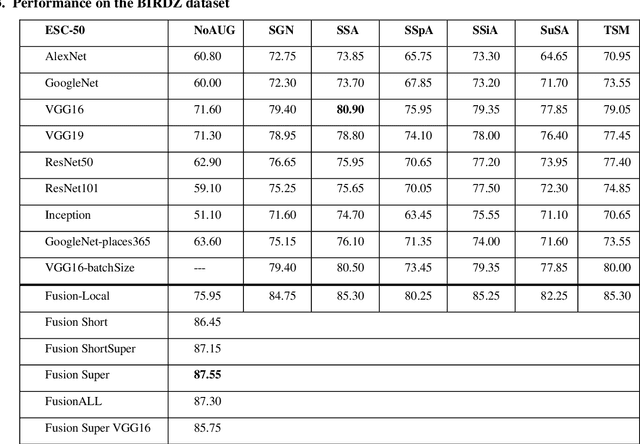
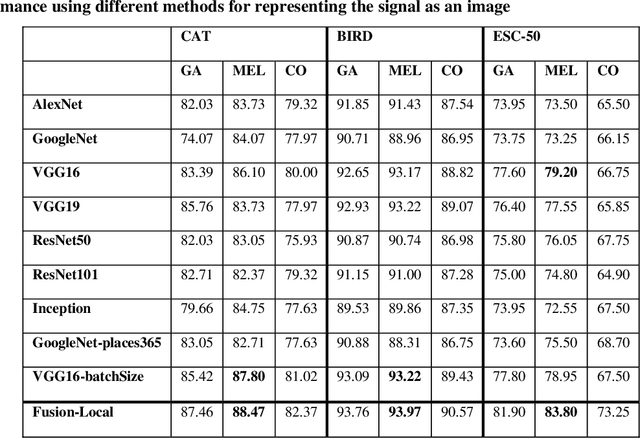
Abstract:In this paper, ensembles of classifiers that exploit several data augmentation techniques and four signal representations for training Convolutional Neural Networks (CNNs) for audio classification are presented and tested on three freely available audio classification datasets: i) bird calls, ii) cat sounds, and iii) the Environmental Sound Classification dataset. The best performing ensembles combining data augmentation techniques with different signal representations are compared and shown to outperform the best methods reported in the literature on these datasets. The approach proposed here obtains state-of-the-art results in the widely used ESC-50 dataset. To the best of our knowledge, this is the most extensive study investigating ensembles of CNNs for audio classification. Results demonstrate not only that CNNs can be trained for audio classification but also that their fusion using different techniques works better than the stand-alone classifiers.
iProStruct2D: Identifying protein structural classes by deep learning via 2D representations
Jun 11, 2019



Abstract:In this paper we address the problem of protein classification starting from a multi-view 2D representation of proteins. From each 3D protein structure, a large set of 2D projections is generated using the protein visualization software Jmol. This set of multi-view 2D representations includes 13 different types of protein visualizations that emphasize specific properties of protein structure (e.g., a backbone visualization that displays the backbone structure of the protein as a trace of the C{\alpha} atom). Each type of representation is used to train a different Convolutional Neural Network (CNN), and the fusion of these CNNs is shown to be able to exploit the diversity of different types of representations to improve classification performance. In addition, several multi-view projections are obtained by uniformly rotating the protein structure around its central X, Y, and Z viewing axes to produce 125 images. This approach can be considered a data augmentation method for improving the performance of the classifier and can be used in both the training and the testing phases. Experimental evaluation of the proposed approach on two datasets demonstrates the strength of the proposed method with respect to the other state-of-the-art approaches. The MATLAB code used in this paper is available at https://github.com/LorisNanni.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge