Gianluca Maguolo
High performing ensemble of convolutional neural networks for insect pest image detection
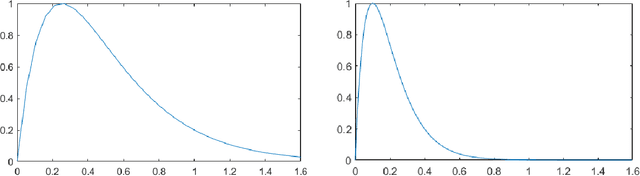
Aug 28, 2021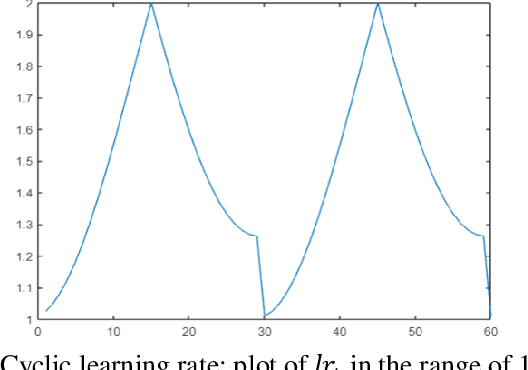
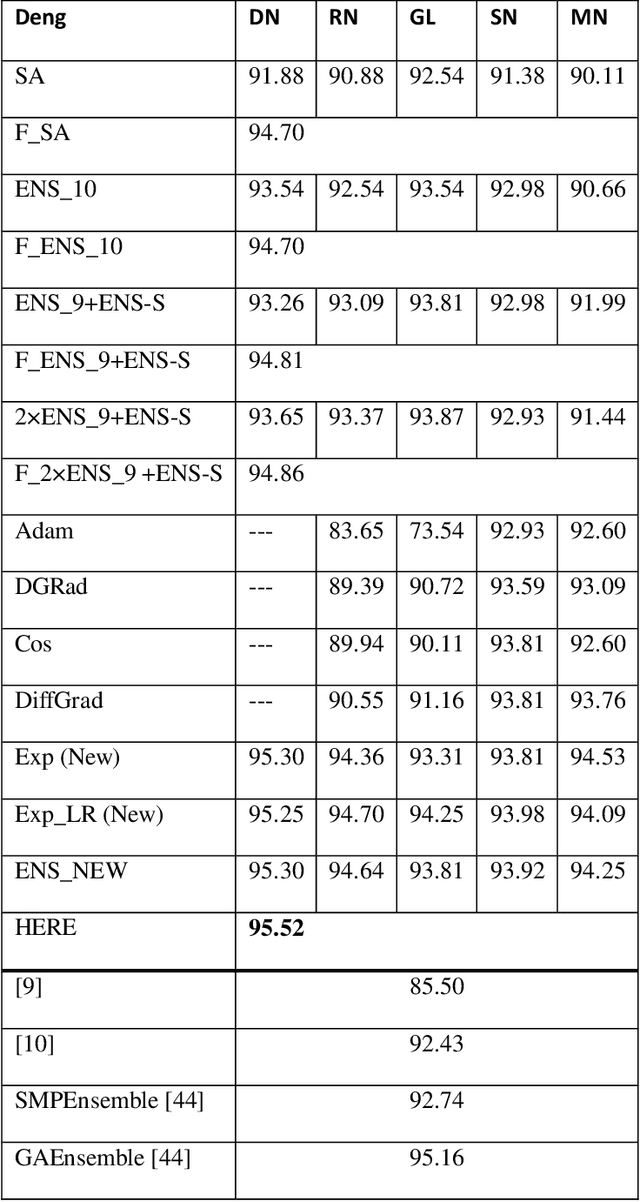

Abstract:Pest infestation is a major cause of crop damage and lost revenues worldwide. Automatic identification of invasive insects would greatly speedup the identification of pests and expedite their removal. In this paper, we generate ensembles of CNNs based on different topologies (ResNet50, GoogleNet, ShuffleNet, MobileNetv2, and DenseNet201) altered by random selection from a simple set of data augmentation methods or optimized with different Adam variants for pest identification. Two new Adam algorithms for deep network optimization based on DGrad are proposed that introduce a scaling factor in the learning rate. Sets of the five CNNs that vary in either data augmentation or the type of Adam optimization were trained on both the Deng (SMALL) and the large IP102 pest data sets. Ensembles were compared and evaluated using three performance indicators. The best performing ensemble, which combined the CNNs using the different augmentation methods and the two new Adam variants proposed here, achieved state of the art on both insect data sets: 95.52% on Deng and 73.46% on IP102, a score on Deng that competed with human expert classifications. Additional tests were performed on data sets for medical imagery classification that further validated the robustness and power of the proposed Adam optimization variants. All MATLAB source code is available at https://github.com/LorisNanni/.
Deep ensembles based on Stochastic Activation Selection for Polyp Segmentation
Apr 07, 2021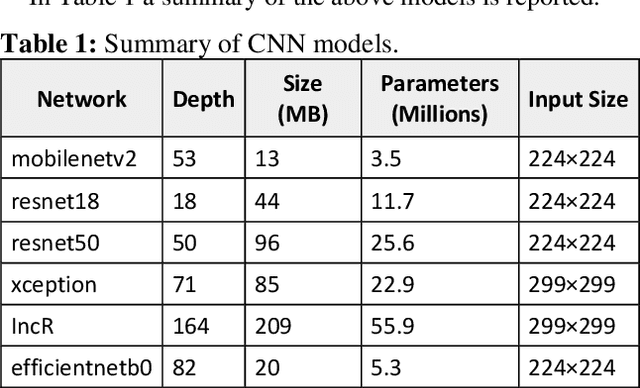
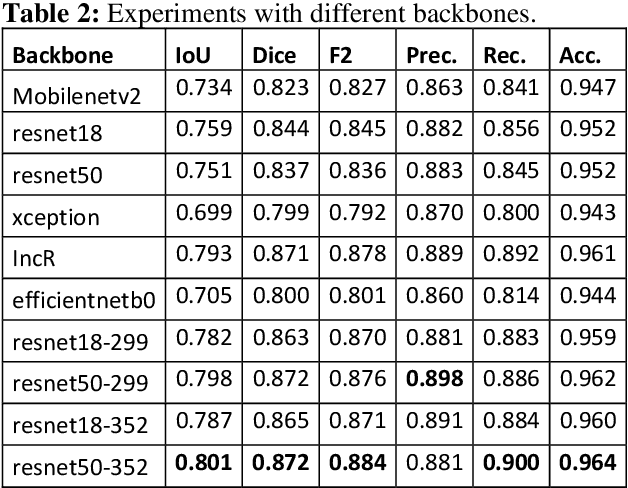
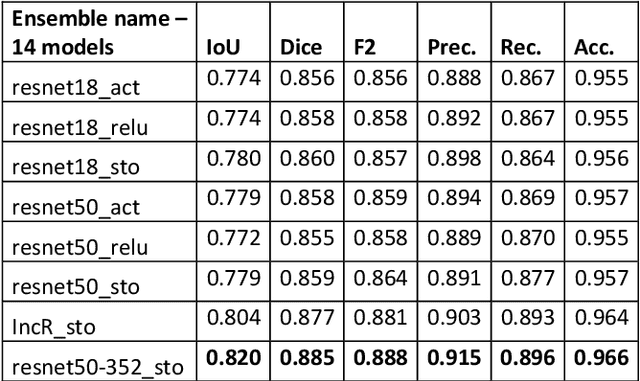
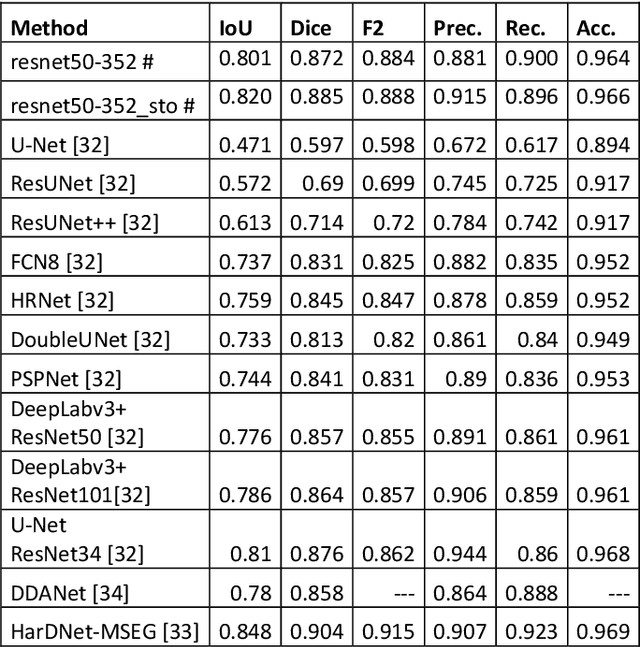
Abstract:Semantic segmentation has a wide array of applications ranging from medical-image analysis, scene understanding, autonomous driving and robotic navigation. This work deals with medical image segmentation and in particular with accurate polyp detection and segmentation during colonoscopy examinations. Several convolutional neural network architectures have been proposed to effectively deal with this task and with the problem of segmenting objects at different scale input. The basic architecture in image segmentation consists of an encoder and a decoder: the first uses convolutional filters to extract features from the image, the second is responsible for generating the final output. In this work, we compare some variant of the DeepLab architecture obtained by varying the decoder backbone. We compare several decoder architectures, including ResNet, Xception, EfficentNet, MobileNet and we perturb their layers by substituting ReLU activation layers with other functions. The resulting methods are used to create deep ensembles which are shown to be very effective. Our experimental evaluations show that our best ensemble produces good segmentation results by achieving high evaluation scores with a dice coefficient of 0.884, and a mean Intersection over Union (mIoU) of 0.818 for the Kvasir-SEG dataset. To improve reproducibility and research efficiency the MATLAB source code used for this research is available at GitHub: https://github.com/LorisNanni.
Comparison of different convolutional neural network activation functions and methods for building ensembles
Apr 02, 2021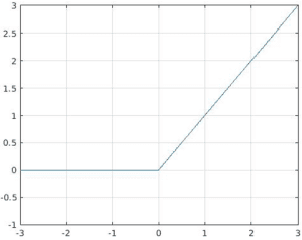
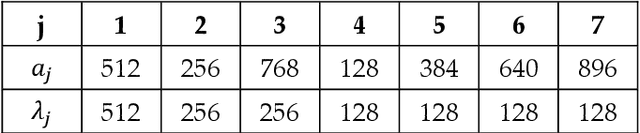
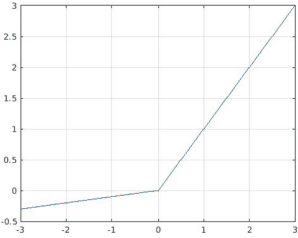
Abstract:Recently, much attention has been devoted to finding highly efficient and powerful activation functions for CNN layers. Because activation functions inject different nonlinearities between layers that affect performance, varying them is one method for building robust ensembles of CNNs. The objective of this study is to examine the performance of CNN ensembles made with different activation functions, including six new ones presented here: 2D Mexican ReLU, TanELU, MeLU+GaLU, Symmetric MeLU, Symmetric GaLU, and Flexible MeLU. The highest performing ensemble was built with CNNs having different activation layers that randomly replaced the standard ReLU. A comprehensive evaluation of the proposed approach was conducted across fifteen biomedical data sets representing various classification tasks. The proposed method was tested on two basic CNN architectures: Vgg16 and ResNet50. Results demonstrate the superiority in performance of this approach. The MATLAB source code for this study will be available at https://github.com/LorisNanni.
Exploiting Adam-like Optimization Algorithms to Improve the Performance of Convolutional Neural Networks
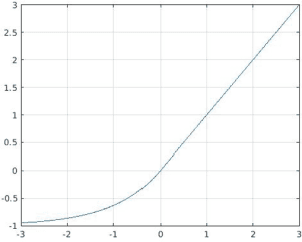
Mar 26, 2021
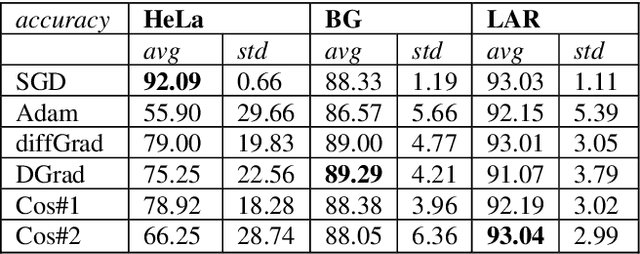
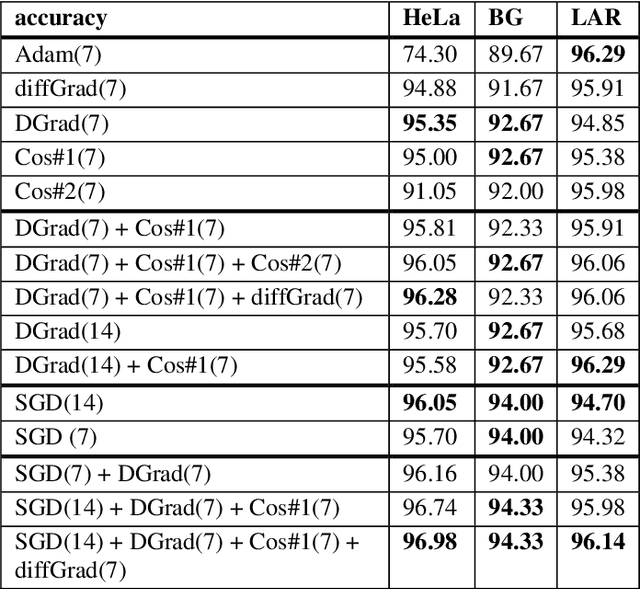
Abstract:Stochastic gradient descent (SGD) is the main approach for training deep networks: it moves towards the optimum of the cost function by iteratively updating the parameters of a model in the direction of the gradient of the loss evaluated on a minibatch. Several variants of SGD have been proposed to make adaptive step sizes for each parameter (adaptive gradient) and take into account the previous updates (momentum). Among several alternative of SGD the most popular are AdaGrad, AdaDelta, RMSProp and Adam which scale coordinates of the gradient by square roots of some form of averaging of the squared coordinates in the past gradients and automatically adjust the learning rate on a parameter basis. In this work, we compare Adam based variants based on the difference between the present and the past gradients, the step size is adjusted for each parameter. We run several tests benchmarking proposed methods using medical image data. The experiments are performed using ResNet50 architecture neural network. Moreover, we have tested ensemble of networks and the fusion with ResNet50 trained with stochastic gradient descent. To combine the set of ResNet50 the simple sum rule has been applied. Proposed ensemble obtains very high performance, it obtains accuracy comparable or better than actual state of the art. To improve reproducibility and research efficiency the MATLAB source code used for this research is available at GitHub: https://github.com/LorisNanni.
Deep learning and hand-crafted features for virus image classification
Dec 13, 2020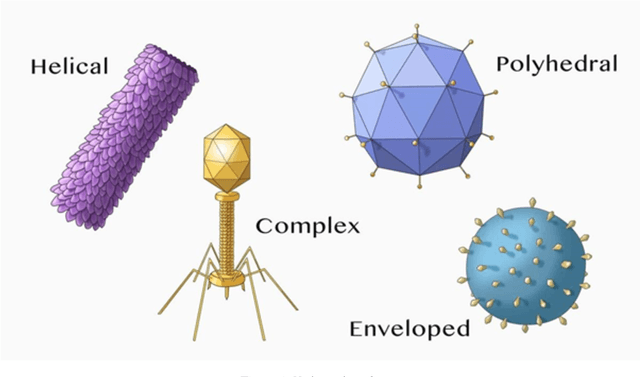

Abstract:In this work, we present an ensemble of descriptors for the classification of transmission electron microscopy images of viruses. We propose to combine handcrafted and deep learning approaches for virus image classification. The set of handcrafted is mainly based on Local Binary Pattern variants, for each descriptor a different Support Vector Machine is trained, then the set of classifiers is combined by sum rule. The deep learning approach is a densenet201 pretrained on ImageNet and then tuned in the virus dataset, the net is used as features extractor for feeding another Support Vector Machine, in particular the last average pooling layer is used as feature extractor. Finally, classifiers trained on handcrafted features and classifier trained on deep learning features are combined by sum rule. The proposed fusion strongly boosts the performance obtained by each stand-alone approach, obtaining state of the art performance.
Comparisons among different stochastic selection of activation layers for convolutional neural networks for healthcare
Nov 24, 2020

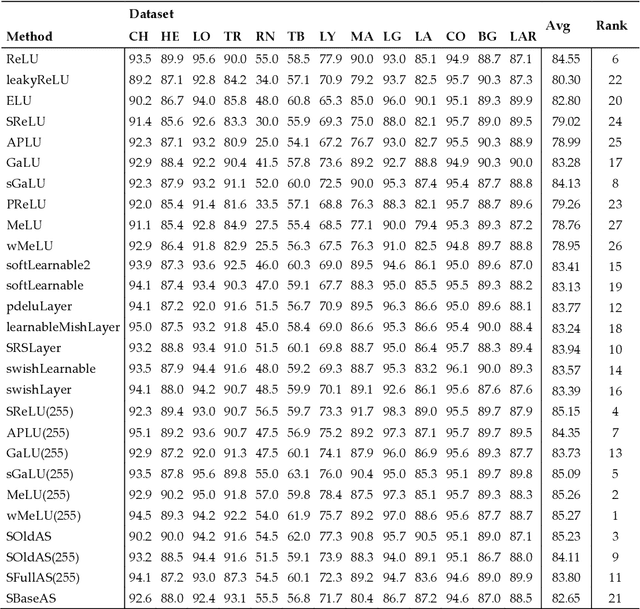
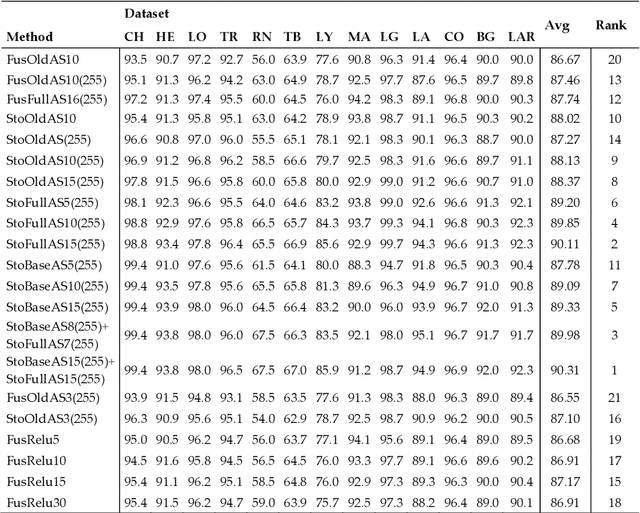
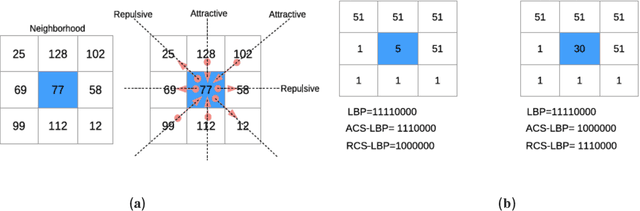
Abstract:Classification of biological images is an important task with crucial application in many fields, such as cell phenotypes recognition, detection of cell organelles and histopathological classification, and it might help in early medical diagnosis, allowing automatic disease classification without the need of a human expert. In this paper we classify biomedical images using ensembles of neural networks. We create this ensemble using a ResNet50 architecture and modifying its activation layers by substituting ReLUs with other functions. We select our activations among the following ones: ReLU, leaky ReLU, Parametric ReLU, ELU, Adaptive Piecewice Linear Unit, S-Shaped ReLU, Swish , Mish, Mexican Linear Unit, Gaussian Linear Unit, Parametric Deformable Linear Unit, Soft Root Sign (SRS) and others. As a baseline, we used an ensemble of neural networks that only use ReLU activations. We tested our networks on several small and medium sized biomedical image datasets. Our results prove that our best ensemble obtains a better performance than the ones of the naive approaches. In order to encourage the reproducibility of this work, the MATLAB code of all the experiments will be shared at https://github.com/LorisNanni.
An Ensemble of Convolutional Neural Networks for Audio Classification
Jul 15, 2020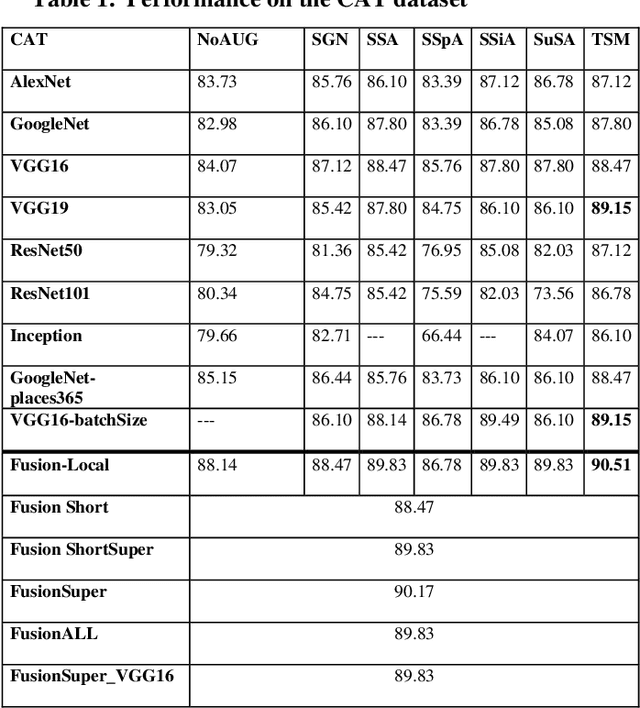

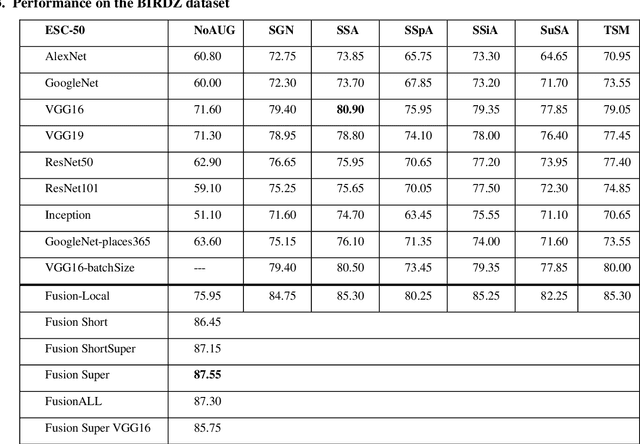
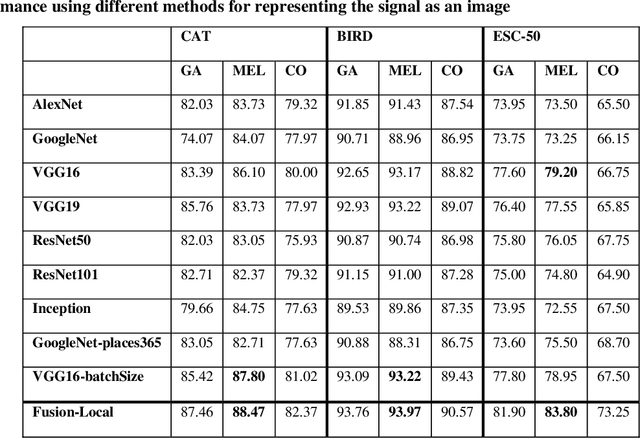
Abstract:In this paper, ensembles of classifiers that exploit several data augmentation techniques and four signal representations for training Convolutional Neural Networks (CNNs) for audio classification are presented and tested on three freely available audio classification datasets: i) bird calls, ii) cat sounds, and iii) the Environmental Sound Classification dataset. The best performing ensembles combining data augmentation techniques with different signal representations are compared and shown to outperform the best methods reported in the literature on these datasets. The approach proposed here obtains state-of-the-art results in the widely used ESC-50 dataset. To the best of our knowledge, this is the most extensive study investigating ensembles of CNNs for audio classification. Results demonstrate not only that CNNs can be trained for audio classification but also that their fusion using different techniques works better than the stand-alone classifiers.
A Critic Evaluation of Methods for COVID-19 Automatic Detection from X-Ray Images
May 19, 2020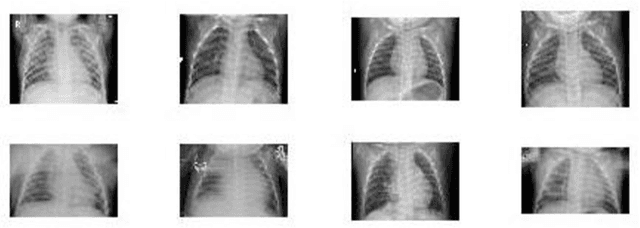

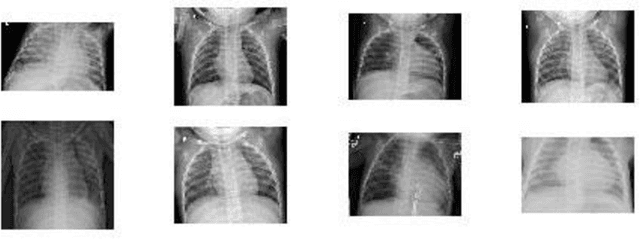
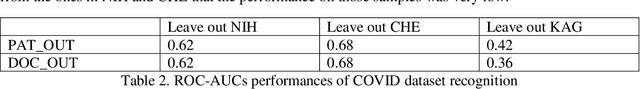
Abstract:In this paper, we compare and evaluate different testing protocols used for automatic COVID-19 diagnosis from X-Ray images in the recent literature. We show that similar results can be obtained using X-Ray images that do not contain most of the lungs. We are able to remove the lungs from the images by turning to black the center of the X-Ray scan and training our classifiers only on the outer part of the images. Hence, we deduce that several testing protocols for the recognition are not fair and that the neural networks are learning patterns in the dataset that are not correlated to the presence of COVID-19. Finally, we show that creating a fair testing protocol is a challenging task, and we provide a method to measure how fair a specific testing protocol is. In the future research we suggest to check the fairness of a testing protocol using our tools and we encourage researchers to look for better techniques than the ones that we propose.
Data augmentation approaches for improving animal audio classification
Dec 16, 2019



Abstract:In this paper we present ensembles of classifiers for automated animal audio classification, exploiting different data augmentation techniques for training Convolutional Neural Networks (CNNs). The specific animal audio classification problems are i) birds and ii) cat sounds, whose datasets are freely available. We train five different CNNs on the original datasets and on their versions augmented by four augmentation protocols, working on the raw audio signals or their representations as spectrograms. We compared our best approaches with the state of the art, showing that we obtain the best recognition rate on the same datasets, without ad hoc parameter optimization. Our study shows that different CNNs can be trained for the purpose of animal audio classification and that their fusion works better than the stand-alone classifiers. To the best of our knowledge this is the largest study on data augmentation for CNNs in animal audio classification audio datasets using the same set of classifiers and parameters. Our MATLAB code is available at https://github.com/LorisNanni .
Audiogmenter: a MATLAB Toolbox for Audio Data Augmentation
Dec 11, 2019



Abstract:Audio data augmentation is a key step in training deep neural networks for solving audio classification tasks. In this paper, we introduce Audiogmenter, a novel audio data augmentation library in MATLAB. We provide 15 different augmentation algorithms for raw audio data and 8 for spectrograms. We integrate the MATLAB built-in audio data augmenter with other methods that proved their effectiveness in literature. To the best of our knowledge, this is the largest MATLAB audio data augmentation library freely available. The toolbox and its documentation can be downloaded at https://github.com/LorisNanni/Audiogmenter
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge