Alberto Bailoni
Proposal-Free Volumetric Instance Segmentation from Latent Single-Instance Masks
Sep 10, 2020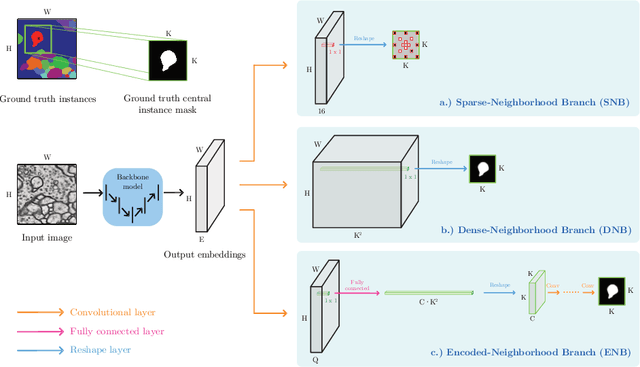
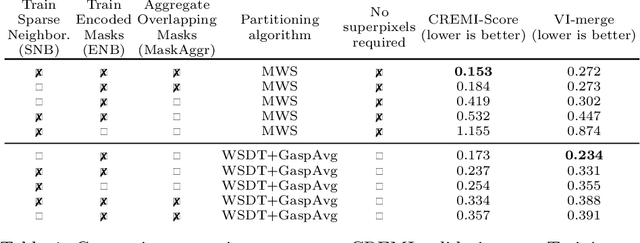
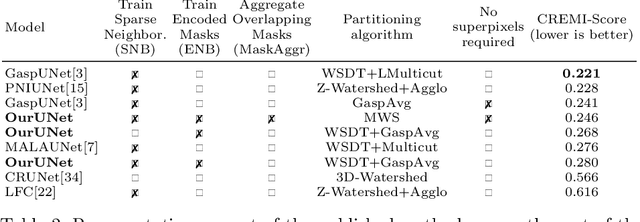
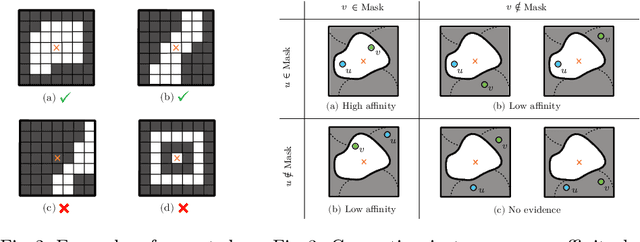
Abstract:This work introduces a new proposal-free instance segmentation method that builds on single-instance segmentation masks predicted across the entire image in a sliding window style. In contrast to related approaches, our method concurrently predicts all masks, one for each pixel, and thus resolves any conflict jointly across the entire image. Specifically, predictions from overlapping masks are combined into edge weights of a signed graph that is subsequently partitioned to obtain all final instances concurrently. The result is a parameter-free method that is strongly robust to noise and prioritizes predictions with the highest consensus across overlapping masks. All masks are decoded from a low dimensional latent representation, which results in great memory savings strictly required for applications to large volumetric images. We test our method on the challenging CREMI 2016 neuron segmentation benchmark where it achieves competitive scores.
The Semantic Mutex Watershed for Efficient Bottom-Up Semantic Instance Segmentation
Dec 29, 2019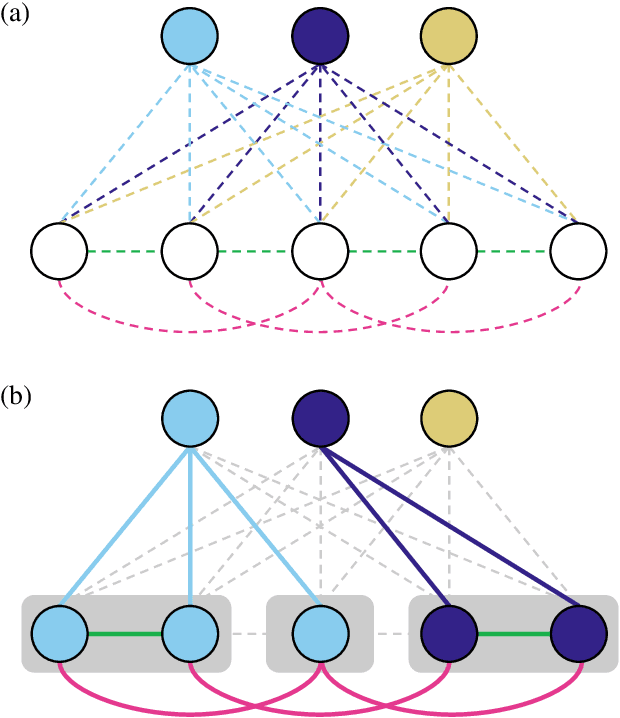
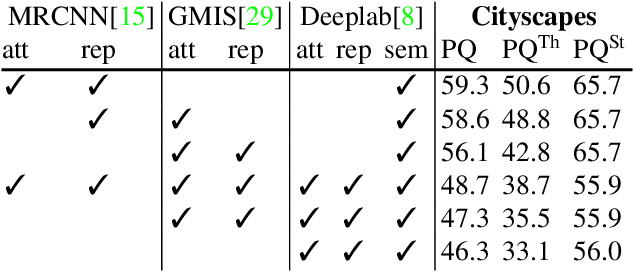

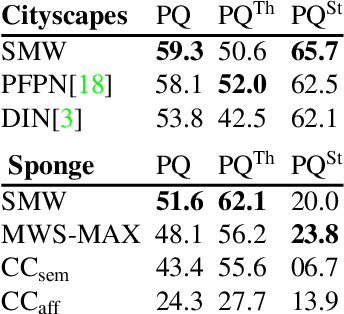
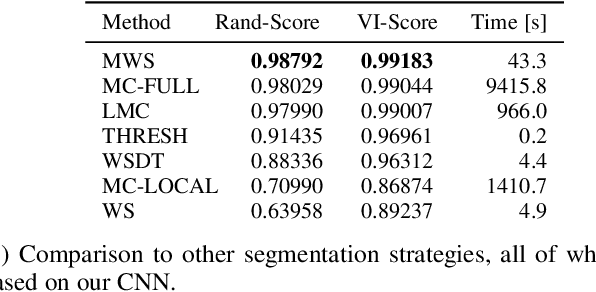
Abstract:Semantic instance segmentation is the task of simultaneously partitioning an image into distinct segments while associating each pixel with a class label. In commonly used pipelines, segmentation and label assignment are solved separately since joint optimization is computationally expensive. We propose a greedy algorithm for joint graph partitioning and labeling derived from the efficient Mutex Watershed partitioning algorithm. It optimizes an objective function closely related to the Symmetric Multiway Cut objective and empirically shows efficient scaling behavior. Due to the algorithm's efficiency it can operate directly on pixels without prior over-segmentation of the image into superpixels. We evaluate the performance on the Cityscapes dataset (2D urban scenes) and on a 3D microscopy volume. In urban scenes, the proposed algorithm combined with current deep neural networks outperforms the strong baseline of `Panoptic Feature Pyramid Networks' by Kirillov et al. (2019). In the 3D electron microscopy images, we show explicitly that our joint formulation outperforms a separate optimization of the partitioning and labeling problems.
DISCo for the CIA: Deep learning, Instance Segmentation, and Correlations for Calcium Imaging Analysis
Aug 22, 2019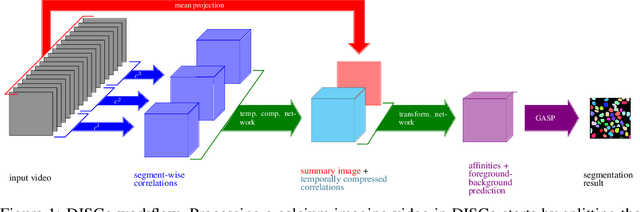

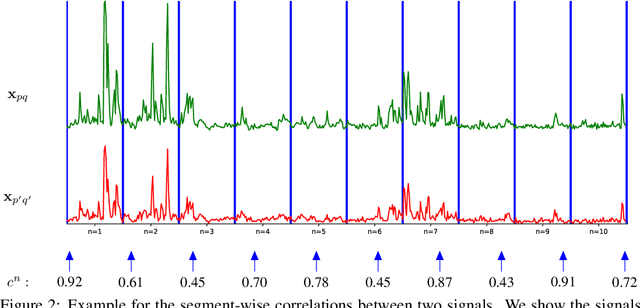
Abstract:Calcium imaging is one of the most important tools in neurophysiology as it enables the observation of neuronal activity for hundreds of cells in parallel and at single-cell resolution. In order to use the data gained with calcium imaging, it is necessary to extract individual cells and their activity from the recordings. Although many sophisticated methods have been proposed, the cell extraction from calcium imaging data can still be prohibitively laborious and require manual annotation and correction. We present DISCo, a novel approach for the cell segmentation in Calcium Imaging Analysis (CIA) that combines the advantages of Deep learning with a state-of-the-art Instance Segmentation algorithm and uses temporal information from the recordings in a computationally efficient way by computing Correlations between pixels.
A Generalized Framework for Agglomerative Clustering of Signed Graphs applied to Instance Segmentation
Jun 27, 2019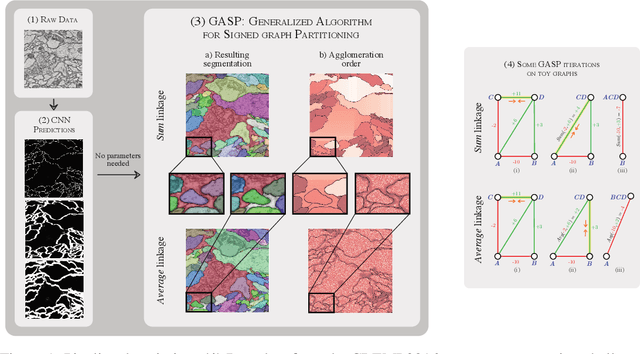
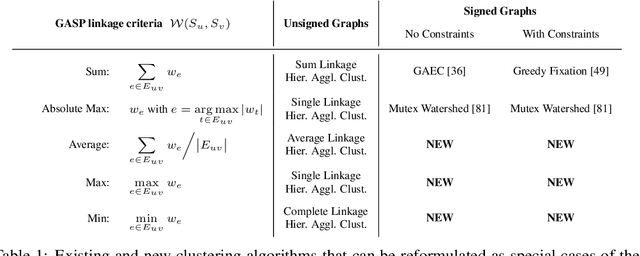
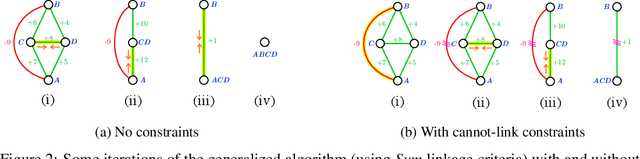
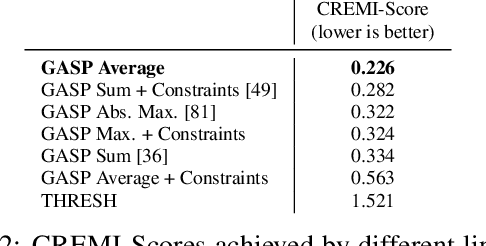
Abstract:We propose a novel theoretical framework that generalizes algorithms for hierarchical agglomerative clustering to weighted graphs with both attractive and repulsive interactions between the nodes. This framework defines GASP, a Generalized Algorithm for Signed graph Partitioning, and allows us to explore many combinations of different linkage criteria and cannot-link constraints. We prove the equivalence of existing clustering methods to some of those combinations, and introduce new algorithms for combinations which have not been studied. An extensive comparison is performed to evaluate properties of the clustering algorithms in the context of instance segmentation in images, including robustness to noise and efficiency. We show how one of the new algorithms proposed in our framework outperforms all previously known agglomerative methods for signed graphs, both on the competitive CREMI 2016 EM segmentation benchmark and on the CityScapes dataset.
The Mutex Watershed and its Objective: Efficient, Parameter-Free Image Partitioning
Apr 25, 2019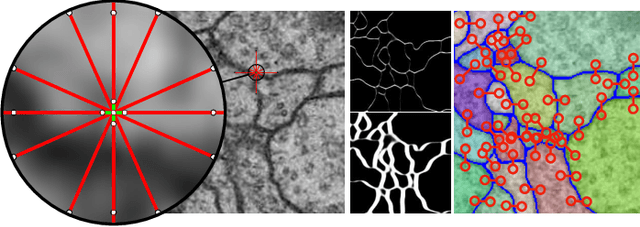
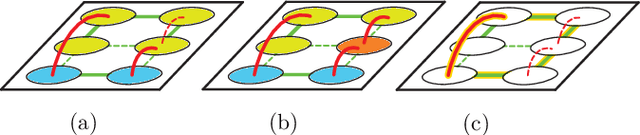
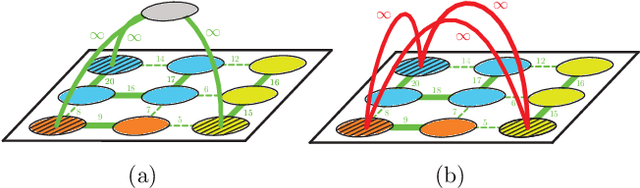
Abstract:Image partitioning, or segmentation without semantics, is the task of decomposing an image into distinct segments, or equivalently to detect closed contours. Most prior work either requires seeds, one per segment; or a threshold; or formulates the task as multicut / correlation clustering, an NP-hard problem. Here, we propose a greedy algorithm for signed graph partitioning, the "Mutex Watershed". Unlike seeded watershed, the algorithm can accommodate not only attractive but also repulsive cues, allowing it to find a previously unspecified number of segments without the need for explicit seeds or a tunable threshold. We also prove that this simple algorithm solves to global optimality an objective function that is intimately related to the multicut / correlation clustering integer linear programming formulation. The algorithm is deterministic, very simple to implement, and has empirically linearithmic complexity. When presented with short-range attractive and long-range repulsive cues from a deep neural network, the Mutex Watershed gives the best results currently known for the competitive ISBI 2012 EM segmentation benchmark.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge