Zhenghao Zeng
Causal Inference for Genomic Data with Multiple Heterogeneous Outcomes
Apr 14, 2024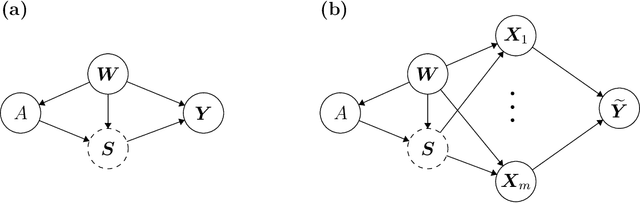
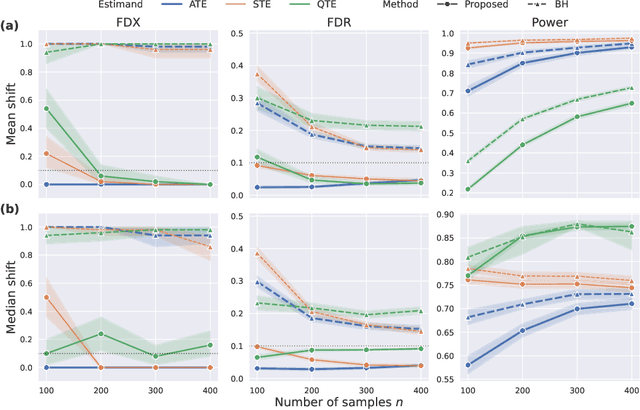
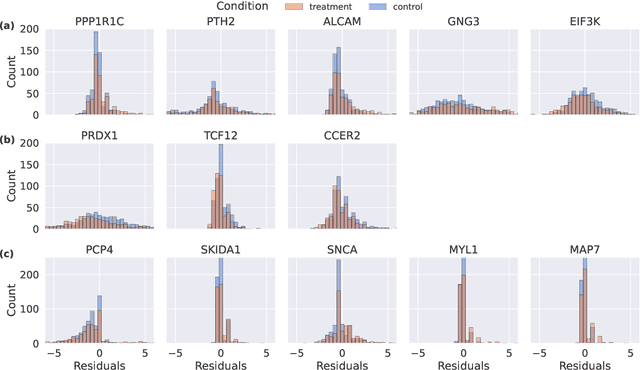
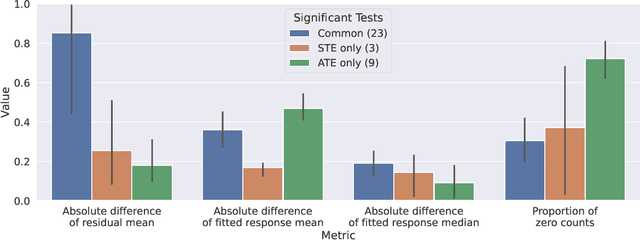
Abstract:With the evolution of single-cell RNA sequencing techniques into a standard approach in genomics, it has become possible to conduct cohort-level causal inferences based on single-cell-level measurements. However, the individual gene expression levels of interest are not directly observable; instead, only repeated proxy measurements from each individual's cells are available, providing a derived outcome to estimate the underlying outcome for each of many genes. In this paper, we propose a generic semiparametric inference framework for doubly robust estimation with multiple derived outcomes, which also encompasses the usual setting of multiple outcomes when the response of each unit is available. To reliably quantify the causal effects of heterogeneous outcomes, we specialize the analysis to the standardized average treatment effects and the quantile treatment effects. Through this, we demonstrate the use of the semiparametric inferential results for doubly robust estimators derived from both Von Mises expansions and estimating equations. A multiple testing procedure based on the Gaussian multiplier bootstrap is tailored for doubly robust estimators to control the false discovery exceedance rate. Applications in single-cell CRISPR perturbation analysis and individual-level differential expression analysis demonstrate the utility of the proposed methods and offer insights into the usage of different estimands for causal inference in genomics.
Continuous Treatment Effects with Surrogate Outcomes
Jan 31, 2024Abstract:In many real-world causal inference applications, the primary outcomes (labels) are often partially missing, especially if they are expensive or difficult to collect. If the missingness depends on covariates (i.e., missingness is not completely at random), analyses based on fully-observed samples alone may be biased. Incorporating surrogates, which are fully observed post-treatment variables related to the primary outcome, can improve estimation in this case. In this paper, we study the role of surrogates in estimating continuous treatment effects and propose a doubly robust method to efficiently incorporate surrogates in the analysis, which uses both labeled and unlabeled data and does not suffer from the above selection bias problem. Importantly, we establish asymptotic normality of the proposed estimator and show possible improvements on the variance compared with methods that solely use labeled data. Extensive simulations show our methods enjoy appealing empirical performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge