Youcheng Li
A Foundational Generative Model for Breast Ultrasound Image Analysis
Jan 12, 2025



Abstract:Foundational models have emerged as powerful tools for addressing various tasks in clinical settings. However, their potential development to breast ultrasound analysis remains untapped. In this paper, we present BUSGen, the first foundational generative model specifically designed for breast ultrasound image analysis. Pretrained on over 3.5 million breast ultrasound images, BUSGen has acquired extensive knowledge of breast structures, pathological features, and clinical variations. With few-shot adaptation, BUSGen can generate repositories of realistic and informative task-specific data, facilitating the development of models for a wide range of downstream tasks. Extensive experiments highlight BUSGen's exceptional adaptability, significantly exceeding real-data-trained foundational models in breast cancer screening, diagnosis, and prognosis. In breast cancer early diagnosis, our approach outperformed all board-certified radiologists (n=9), achieving an average sensitivity improvement of 16.5% (P-value<0.0001). Additionally, we characterized the scaling effect of using generated data which was as effective as the collected real-world data for training diagnostic models. Moreover, extensive experiments demonstrated that our approach improved the generalization ability of downstream models. Importantly, BUSGen protected patient privacy by enabling fully de-identified data sharing, making progress forward in secure medical data utilization. An online demo of BUSGen is available at https://aibus.bio.
Knowledge-driven AI-generated data for accurate and interpretable breast ultrasound diagnoses
Jul 23, 2024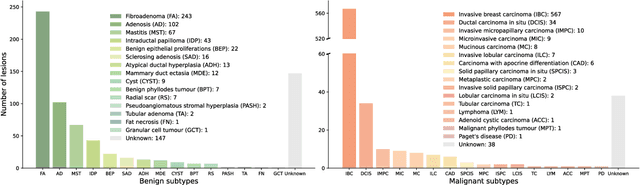
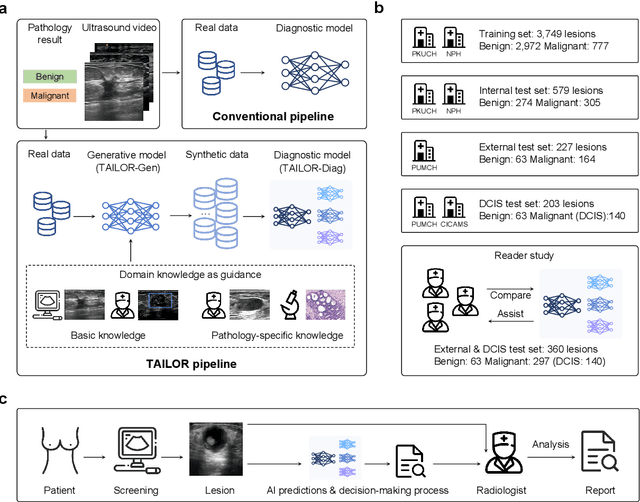
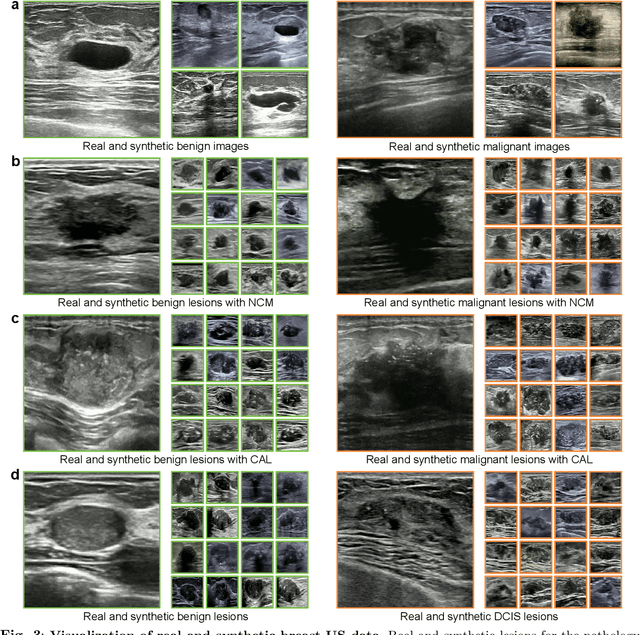
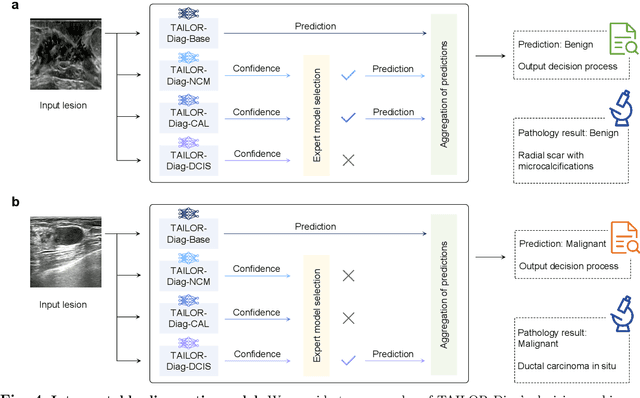
Abstract:Data-driven deep learning models have shown great capabilities to assist radiologists in breast ultrasound (US) diagnoses. However, their effectiveness is limited by the long-tail distribution of training data, which leads to inaccuracies in rare cases. In this study, we address a long-standing challenge of improving the diagnostic model performance on rare cases using long-tailed data. Specifically, we introduce a pipeline, TAILOR, that builds a knowledge-driven generative model to produce tailored synthetic data. The generative model, using 3,749 lesions as source data, can generate millions of breast-US images, especially for error-prone rare cases. The generated data can be further used to build a diagnostic model for accurate and interpretable diagnoses. In the prospective external evaluation, our diagnostic model outperforms the average performance of nine radiologists by 33.5% in specificity with the same sensitivity, improving their performance by providing predictions with an interpretable decision-making process. Moreover, on ductal carcinoma in situ (DCIS), our diagnostic model outperforms all radiologists by a large margin, with only 34 DCIS lesions in the source data. We believe that TAILOR can potentially be extended to various diseases and imaging modalities.
Mining Negative Temporal Contexts For False Positive Suppression In Real-Time Ultrasound Lesion Detection
May 29, 2023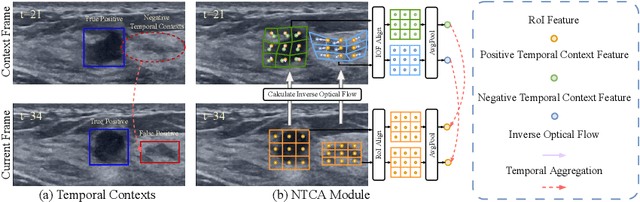
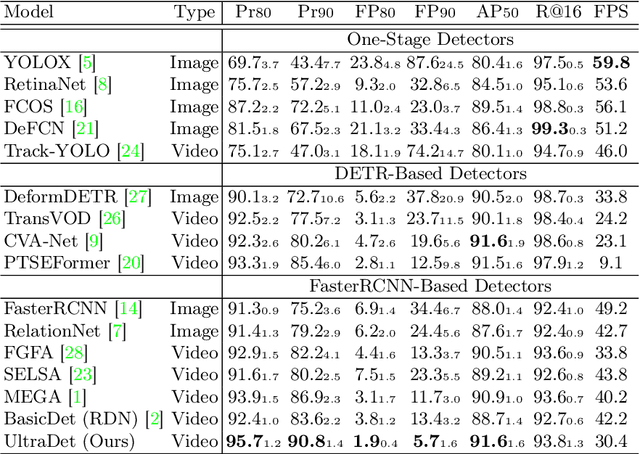
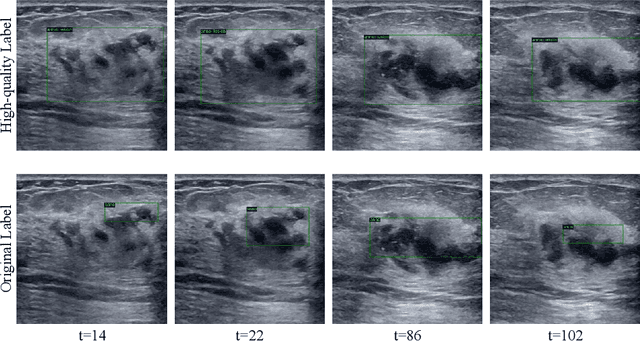

Abstract:During ultrasonic scanning processes, real-time lesion detection can assist radiologists in accurate cancer diagnosis. However, this essential task remains challenging and underexplored. General-purpose real-time object detection models can mistakenly report obvious false positives (FPs) when applied to ultrasound videos, potentially misleading junior radiologists. One key issue is their failure to utilize negative symptoms in previous frames, denoted as negative temporal contexts (NTC). To address this issue, we propose to extract contexts from previous frames, including NTC, with the guidance of inverse optical flow. By aggregating extracted contexts, we endow the model with the ability to suppress FPs by leveraging NTC. We call the resulting model UltraDet. The proposed UltraDet demonstrates significant improvement over previous state-of-the-arts and achieves real-time inference speed. To facilitate future research, we will release the code, checkpoints, and high-quality labels of the CVA-BUS dataset used in our experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge