Yizhong Liu
Learning Representation and Synergy Invariances: A Povable Framework for Generalized Multimodal Face Anti-Spoofing
Nov 18, 2025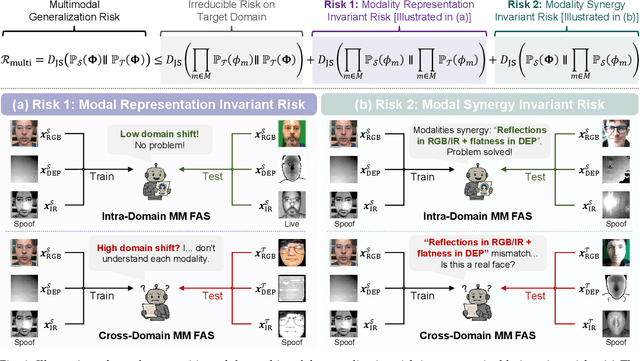
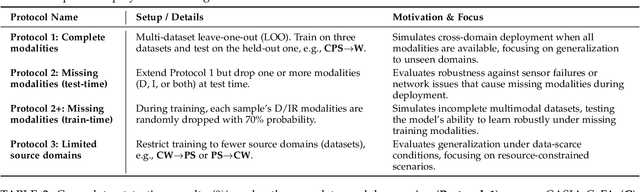
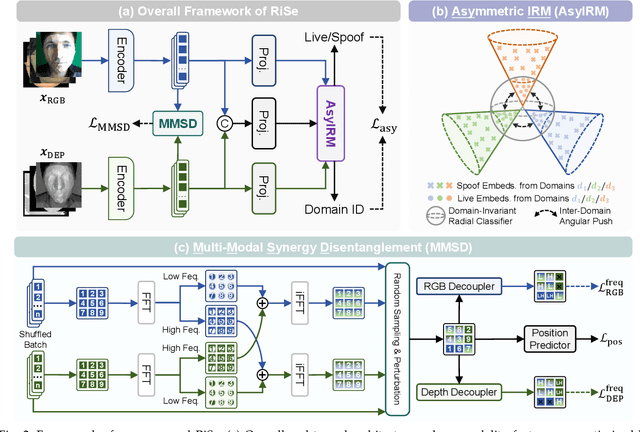
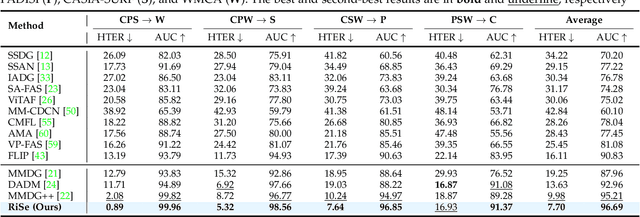
Abstract:Multimodal Face Anti-Spoofing (FAS) methods, which integrate multiple visual modalities, often suffer even more severe performance degradation than unimodal FAS when deployed in unseen domains. This is mainly due to two overlooked risks that affect cross-domain multimodal generalization. The first is the modal representation invariant risk, i.e., whether representations remain generalizable under domain shift. We theoretically show that the inherent class asymmetry in FAS (diverse spoofs vs. compact reals) enlarges the upper bound of generalization error, and this effect is further amplified in multimodal settings. The second is the modal synergy invariant risk, where models overfit to domain-specific inter-modal correlations. Such spurious synergy cannot generalize to unseen attacks in target domains, leading to performance drops. To solve these issues, we propose a provable framework, namely Multimodal Representation and Synergy Invariance Learning (RiSe). For representation risk, RiSe introduces Asymmetric Invariant Risk Minimization (AsyIRM), which learns an invariant spherical decision boundary in radial space to fit asymmetric distributions, while preserving domain cues in angular space. For synergy risk, RiSe employs Multimodal Synergy Disentanglement (MMSD), a self-supervised task enhancing intrinsic, generalizable modal features via cross-sample mixing and disentanglement. Theoretical analysis and experiments verify RiSe, which achieves state-of-the-art cross-domain performance.
Sparsification Under Siege: Defending Against Poisoning Attacks in Communication-Efficient Federated Learning
Apr 30, 2025



Abstract:Federated Learning (FL) enables collaborative model training across distributed clients while preserving data privacy, yet it faces significant challenges in communication efficiency and vulnerability to poisoning attacks. While sparsification techniques mitigate communication overhead by transmitting only critical model parameters, they inadvertently amplify security risks: adversarial clients can exploit sparse updates to evade detection and degrade model performance. Existing defense mechanisms, designed for standard FL communication scenarios, are ineffective in addressing these vulnerabilities within sparsified FL. To bridge this gap, we propose FLARE, a novel federated learning framework that integrates sparse index mask inspection and model update sign similarity analysis to detect and mitigate poisoning attacks in sparsified FL. Extensive experiments across multiple datasets and adversarial scenarios demonstrate that FLARE significantly outperforms existing defense strategies, effectively securing sparsified FL against poisoning attacks while maintaining communication efficiency.
Safeguarding Medical Image Segmentation Datasets against Unauthorized Training via Contour- and Texture-Aware Perturbations
Mar 21, 2024Abstract:The widespread availability of publicly accessible medical images has significantly propelled advancements in various research and clinical fields. Nonetheless, concerns regarding unauthorized training of AI systems for commercial purposes and the duties of patient privacy protection have led numerous institutions to hesitate to share their images. This is particularly true for medical image segmentation (MIS) datasets, where the processes of collection and fine-grained annotation are time-intensive and laborious. Recently, Unlearnable Examples (UEs) methods have shown the potential to protect images by adding invisible shortcuts. These shortcuts can prevent unauthorized deep neural networks from generalizing. However, existing UEs are designed for natural image classification and fail to protect MIS datasets imperceptibly as their protective perturbations are less learnable than important prior knowledge in MIS, e.g., contour and texture features. To this end, we propose an Unlearnable Medical image generation method, termed UMed. UMed integrates the prior knowledge of MIS by injecting contour- and texture-aware perturbations to protect images. Given that our target is to only poison features critical to MIS, UMed requires only minimal perturbations within the ROI and its contour to achieve greater imperceptibility (average PSNR is 50.03) and protective performance (clean average DSC degrades from 82.18% to 6.80%).
Suppress and Rebalance: Towards Generalized Multi-Modal Face Anti-Spoofing
Mar 05, 2024



Abstract:Face Anti-Spoofing (FAS) is crucial for securing face recognition systems against presentation attacks. With advancements in sensor manufacture and multi-modal learning techniques, many multi-modal FAS approaches have emerged. However, they face challenges in generalizing to unseen attacks and deployment conditions. These challenges arise from (1) modality unreliability, where some modality sensors like depth and infrared undergo significant domain shifts in varying environments, leading to the spread of unreliable information during cross-modal feature fusion, and (2) modality imbalance, where training overly relies on a dominant modality hinders the convergence of others, reducing effectiveness against attack types that are indistinguishable sorely using the dominant modality. To address modality unreliability, we propose the Uncertainty-Guided Cross-Adapter (U-Adapter) to recognize unreliably detected regions within each modality and suppress the impact of unreliable regions on other modalities. For modality imbalance, we propose a Rebalanced Modality Gradient Modulation (ReGrad) strategy to rebalance the convergence speed of all modalities by adaptively adjusting their gradients. Besides, we provide the first large-scale benchmark for evaluating multi-modal FAS performance under domain generalization scenarios. Extensive experiments demonstrate that our method outperforms state-of-the-art methods. Source code and protocols will be released on https://github.com/OMGGGGG/mmdg.
Biomedical Image Splicing Detection using Uncertainty-Guided Refinement
Sep 28, 2023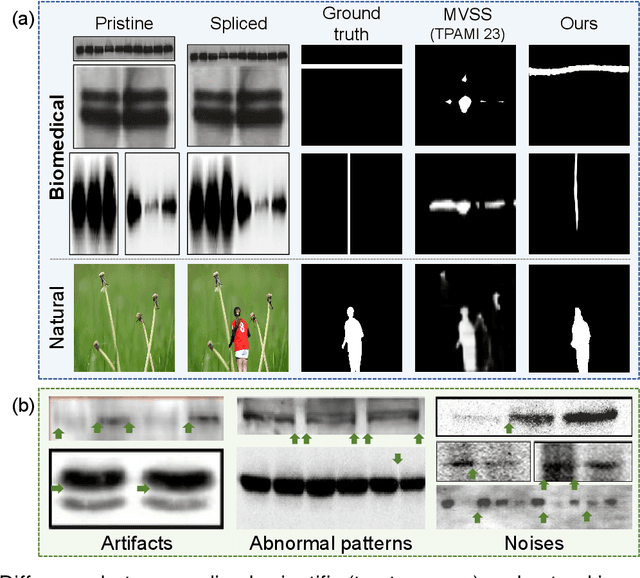
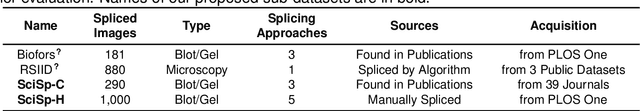
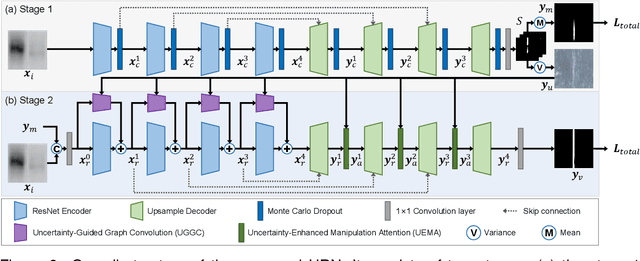
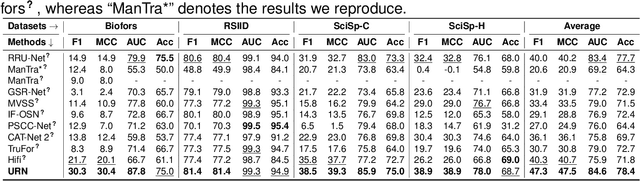
Abstract:Recently, a surge in biomedical academic publications suspected of image manipulation has led to numerous retractions, turning biomedical image forensics into a research hotspot. While manipulation detectors are concerning, the specific detection of splicing traces in biomedical images remains underexplored. The disruptive factors within biomedical images, such as artifacts, abnormal patterns, and noises, show misleading features like the splicing traces, greatly increasing the challenge for this task. Moreover, the scarcity of high-quality spliced biomedical images also limits potential advancements in this field. In this work, we propose an Uncertainty-guided Refinement Network (URN) to mitigate the effects of these disruptive factors. Our URN can explicitly suppress the propagation of unreliable information flow caused by disruptive factors among regions, thereby obtaining robust features. Moreover, URN enables a concentration on the refinement of uncertainly predicted regions during the decoding phase. Besides, we construct a dataset for Biomedical image Splicing (BioSp) detection, which consists of 1,290 spliced images. Compared with existing datasets, BioSp comprises the largest number of spliced images and the most diverse sources. Comprehensive experiments on three benchmark datasets demonstrate the superiority of the proposed method. Meanwhile, we verify the generalizability of URN when against cross-dataset domain shifts and its robustness to resist post-processing approaches. Our BioSp dataset will be released upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge