Yanming Sun
Beyond Simple Fusion: Adaptive Gated Fusion for Robust Multimodal Sentiment Analysis
Oct 02, 2025



Abstract:Multimodal sentiment analysis (MSA) leverages information fusion from diverse modalities (e.g., text, audio, visual) to enhance sentiment prediction. However, simple fusion techniques often fail to account for variations in modality quality, such as those that are noisy, missing, or semantically conflicting. This oversight leads to suboptimal performance, especially in discerning subtle emotional nuances. To mitigate this limitation, we introduce a simple yet efficient \textbf{A}daptive \textbf{G}ated \textbf{F}usion \textbf{N}etwork that adaptively adjusts feature weights via a dual gate fusion mechanism based on information entropy and modality importance. This mechanism mitigates the influence of noisy modalities and prioritizes informative cues following unimodal encoding and cross-modal interaction. Experiments on CMU-MOSI and CMU-MOSEI show that AGFN significantly outperforms strong baselines in accuracy, effectively discerning subtle emotions with robust performance. Visualization analysis of feature representations demonstrates that AGFN enhances generalization by learning from a broader feature distribution, achieved by reducing the correlation between feature location and prediction error, thereby decreasing reliance on specific locations and creating more robust multimodal feature representations.
Exposing the Cracks: Vulnerabilities of Retrieval-Augmented LLM-based Machine Translation
Oct 01, 2025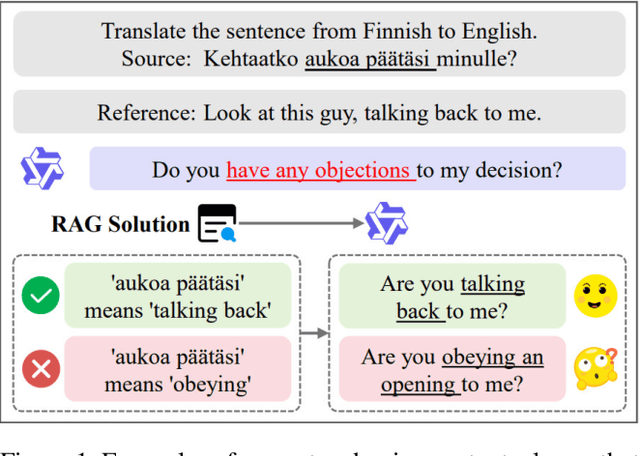
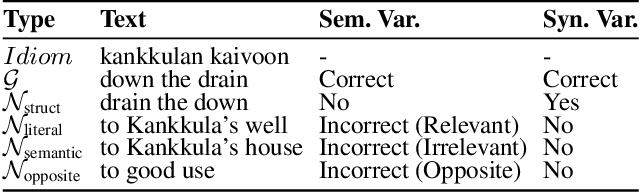
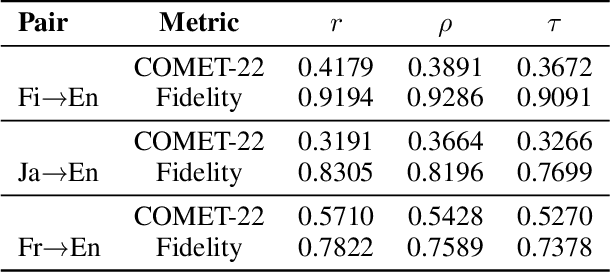
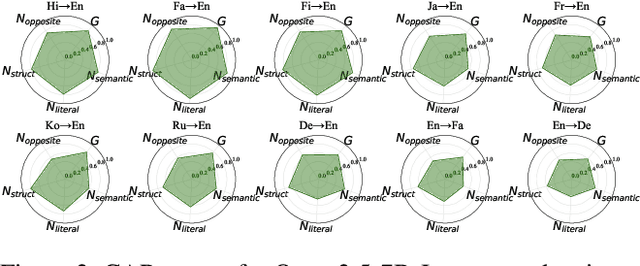
Abstract:\textbf{RE}trieval-\textbf{A}ugmented \textbf{L}LM-based \textbf{M}achine \textbf{T}ranslation (REAL-MT) shows promise for knowledge-intensive tasks like idiomatic translation, but its reliability under noisy retrieval contexts remains poorly understood despite this being a common challenge in real-world deployment. To address this gap, we propose a noise synthesis framework and new metrics to evaluate the robustness of REAL-MT systematically. Using this framework, we instantiate REAL-MT with Qwen-series models, including standard LLMs and large reasoning models (LRMs) with enhanced reasoning, and evaluate their performance on idiomatic translation across high-, medium-, and low-resource language pairs under synthesized noise. Our results show that low-resource language pairs, which rely more heavily on retrieved context, degrade more severely under noise than high-resource ones and often produce nonsensical translations. Although LRMs possess enhanced reasoning capabilities, they show no improvement in error correction and are even more susceptible to noise, tending to rationalize incorrect contexts. We find that this stems from an attention shift away from the source idiom to noisy content, while confidence increases despite declining accuracy, indicating poor calibration. To mitigate these issues, we investigate training-free and fine-tuning strategies, which improve robustness at the cost of performance in clean contexts, revealing a fundamental trade-off. Our findings highlight the limitations of current approaches, underscoring the need for self-verifying integration mechanisms.
Brain Tumor Detection Based on a Novel and High-Quality Prediction of the Tumor Pixel Distributions
Aug 14, 2023Abstract:In this paper, we propose a system to detect brain tumor in 3D MRI brain scans of Flair modality. It performs 2 functions: (a) predicting gray-level and locational distributions of the pixels in the tumor regions and (b) generating tumor mask in pixel-wise precision. To facilitate 3D data analysis and processing, we introduced a 2D histogram presentation that comprehends the gray-level distribution and pixel-location distribution of a 3D object. In the proposed system, particular 2D histograms, in which tumor-related feature data get concentrated, are established by exploiting the left-right asymmetry of a brain structure. A modulation function is generated from the input data of each patient case and applied to the 2D histograms to attenuate the element irrelevant to the tumor regions. The prediction of the tumor pixel distribution is done in 3 steps, on the axial, coronal and sagittal slice series, respectively. In each step, the prediction result helps to identify/remove tumor-free slices, increasing the tumor information density in the remaining data to be applied to the next step. After the 3-step removal, the 3D input is reduced to a minimum bounding box of the tumor region. It is used to finalize the prediction and then transformed into a 3D tumor mask, by means of gray level thresholding and low-pass-based morphological operations. The final prediction result is used to determine the critical threshold. The proposed system has been tested extensively with the data of more than one thousand patient cases in the datasets of BraTS 2018~21. The test results demonstrate that the predicted 2D histograms have a high degree of similarity with the true ones. The system delivers also very good tumor detection results, comparable to those of state-of-the-art CNN systems with mono-modality inputs, which is achieved at an extremely low computation cost and no need for training.
TempoSum: Evaluating the Temporal Generalization of Abstractive Summarization
May 03, 2023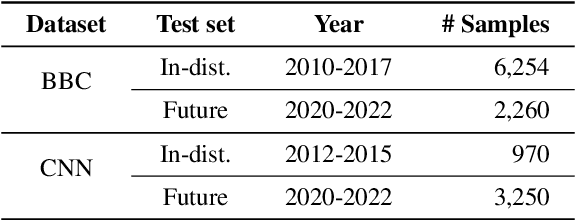
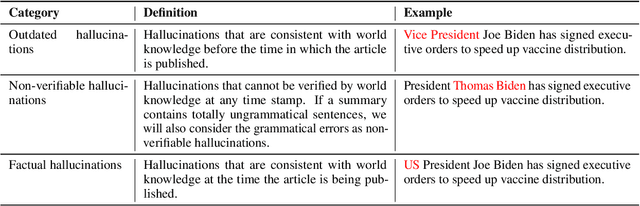
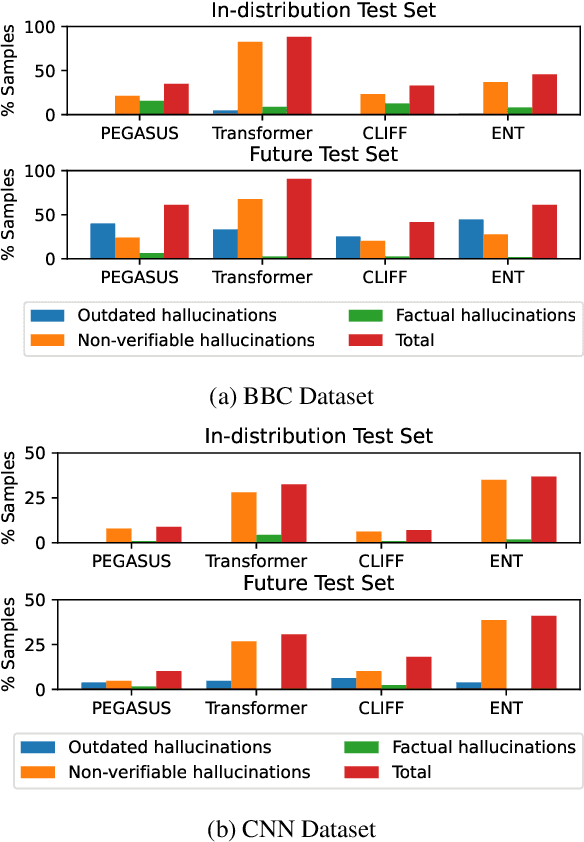
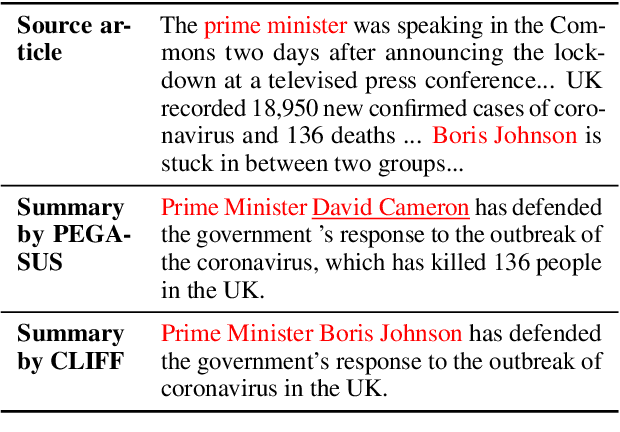
Abstract:Recent pre-trained language models (PLMs) achieve promising results in existing abstractive summarization datasets. However, existing summarization benchmarks overlap in time with the standard pre-training corpora and finetuning datasets. Hence, the strong performance of PLMs may rely on the parametric knowledge that is memorized during pre-training and fine-tuning. Moreover, the knowledge memorized by PLMs may quickly become outdated, which affects the generalization performance of PLMs on future data. In this work, we propose TempoSum, a novel benchmark that contains data samples from 2010 to 2022, to understand the temporal generalization ability of abstractive summarization models. Through extensive human evaluation, we show that parametric knowledge stored in summarization models significantly affects the faithfulness of the generated summaries on future data. Moreover, existing faithfulness enhancement methods cannot reliably improve the faithfulness of summarization models on future data. Finally, we discuss several recommendations to the research community on how to evaluate and improve the temporal generalization capability of text summarization models.
A Computation-Efficient CNN System for High-Quality Brain Tumor Segmentation
Aug 07, 2020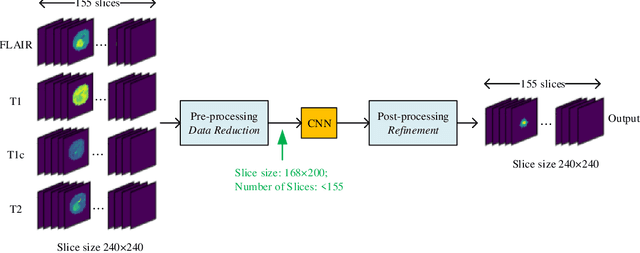
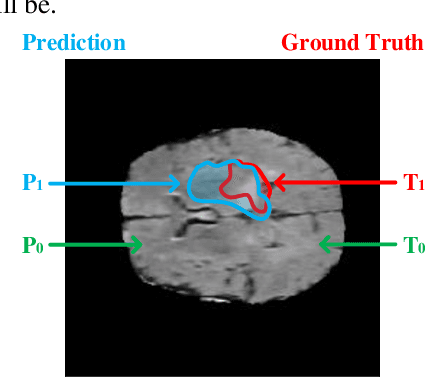
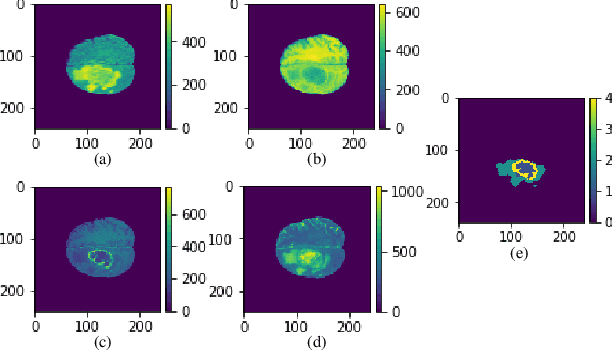
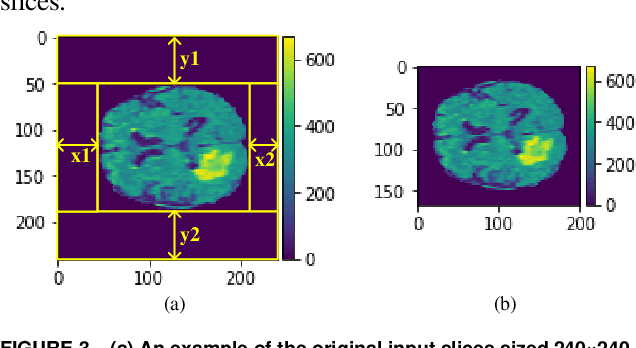
Abstract:In this paper, a Convolutional Neural Network (CNN) system is proposed for brain tumor segmentation. The system consists of three parts, a pre-processing block to reduce the data volume, an application-specific CNN(ASCNN) to segment tumor areas precisely, and a refinement block to detect/remove false positive pixels. The CNN, designed specifically for the task, has 7 convolution layers, 16 channels per layer, requiring only 11716 parameters. The convolutions combined with max-pooling in the first half of the CNN are performed to localize tumor areas. Two convolution modes, namely depthwise convolution and standard convolution, are performed in parallel in the first 2 layers to extract elementary features efficiently. For a fine classification of pixel-wise precision in the second half of the CNN, the feature maps are modulated by adding the individually weighted local feature maps generated in the first half of the CNN. The performance of the proposed system has been evaluated by an online platform with dataset of Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) 2018. Requiring a very low computation volume, the proposed system delivers a high segmentation quality indicated by its average Dice scores of 0.75, 0.88 and 0.76 for enhancing tumor, whole tumor and tumor core, respectively, and also by the median Dice scores of 0.85, 0.92, and 0.86. The consistency in system performance has also been measured, demonstrating that the system is able to reproduce almost the same output to the same input after retraining. The simple structure of the proposed system facilitates its implementation in computation restricted environment, and a wide range of applications can thus be expected.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge