Chunyan Wang
Diffusion-Guided Knowledge Distillation for Weakly-Supervised Low-Light Semantic Segmentation
Jul 10, 2025Abstract:Weakly-supervised semantic segmentation aims to assign category labels to each pixel using weak annotations, significantly reducing manual annotation costs. Although existing methods have achieved remarkable progress in well-lit scenarios, their performance significantly degrades in low-light environments due to two fundamental limitations: severe image quality degradation (e.g., low contrast, noise, and color distortion) and the inherent constraints of weak supervision. These factors collectively lead to unreliable class activation maps and semantically ambiguous pseudo-labels, ultimately compromising the model's ability to learn discriminative feature representations. To address these problems, we propose Diffusion-Guided Knowledge Distillation for Weakly-Supervised Low-light Semantic Segmentation (DGKD-WLSS), a novel framework that synergistically combines Diffusion-Guided Knowledge Distillation (DGKD) with Depth-Guided Feature Fusion (DGF2). DGKD aligns normal-light and low-light features via diffusion-based denoising and knowledge distillation, while DGF2 integrates depth maps as illumination-invariant geometric priors to enhance structural feature learning. Extensive experiments demonstrate the effectiveness of DGKD-WLSS, which achieves state-of-the-art performance in weakly supervised semantic segmentation tasks under low-light conditions. The source codes have been released at:https://github.com/ChunyanWang1/DGKD-WLSS.
Potential of large language model-powered nudges for promoting daily water and energy conservation
Mar 14, 2025Abstract:The increasing amount of pressure related to water and energy shortages has increased the urgency of cultivating individual conservation behaviors. While the concept of nudging, i.e., providing usage-based feedback, has shown promise in encouraging conservation behaviors, its efficacy is often constrained by the lack of targeted and actionable content. This study investigates the impact of the use of large language models (LLMs) to provide tailored conservation suggestions for conservation intentions and their rationale. Through a survey experiment with 1,515 university participants, we compare three virtual nudging scenarios: no nudging, traditional nudging with usage statistics, and LLM-powered nudging with usage statistics and personalized conservation suggestions. The results of statistical analyses and causal forest modeling reveal that nudging led to an increase in conservation intentions among 86.9%-98.0% of the participants. LLM-powered nudging achieved a maximum increase of 18.0% in conservation intentions, surpassing traditional nudging by 88.6%. Furthermore, structural equation modeling results reveal that exposure to LLM-powered nudges enhances self-efficacy and outcome expectations while diminishing dependence on social norms, thereby increasing intrinsic motivation to conserve. These findings highlight the transformative potential of LLMs in promoting individual water and energy conservation, representing a new frontier in the design of sustainable behavioral interventions and resource management.
Boosting Weakly-Supervised Image Segmentation via Representation, Transform, and Compensator
Sep 02, 2023Abstract:Weakly-supervised image segmentation (WSIS) is a critical task in computer vision that relies on image-level class labels. Multi-stage training procedures have been widely used in existing WSIS approaches to obtain high-quality pseudo-masks as ground-truth, resulting in significant progress. However, single-stage WSIS methods have recently gained attention due to their potential for simplifying training procedures, despite often suffering from low-quality pseudo-masks that limit their practical applications. To address this issue, we propose a novel single-stage WSIS method that utilizes a siamese network with contrastive learning to improve the quality of class activation maps (CAMs) and achieve a self-refinement process. Our approach employs a cross-representation refinement method that expands reliable object regions by utilizing different feature representations from the backbone. Additionally, we introduce a cross-transform regularization module that learns robust class prototypes for contrastive learning and captures global context information to feed back rough CAMs, thereby improving the quality of CAMs. Our final high-quality CAMs are used as pseudo-masks to supervise the segmentation result. Experimental results on the PASCAL VOC 2012 dataset demonstrate that our method significantly outperforms other state-of-the-art methods, achieving 67.2% and 68.76% mIoU on PASCAL VOC 2012 val set and test set, respectively. Furthermore, our method has been extended to weakly supervised object localization task, and experimental results demonstrate that our method continues to achieve very competitive results.
Brain Tumor Detection Based on a Novel and High-Quality Prediction of the Tumor Pixel Distributions
Aug 14, 2023Abstract:In this paper, we propose a system to detect brain tumor in 3D MRI brain scans of Flair modality. It performs 2 functions: (a) predicting gray-level and locational distributions of the pixels in the tumor regions and (b) generating tumor mask in pixel-wise precision. To facilitate 3D data analysis and processing, we introduced a 2D histogram presentation that comprehends the gray-level distribution and pixel-location distribution of a 3D object. In the proposed system, particular 2D histograms, in which tumor-related feature data get concentrated, are established by exploiting the left-right asymmetry of a brain structure. A modulation function is generated from the input data of each patient case and applied to the 2D histograms to attenuate the element irrelevant to the tumor regions. The prediction of the tumor pixel distribution is done in 3 steps, on the axial, coronal and sagittal slice series, respectively. In each step, the prediction result helps to identify/remove tumor-free slices, increasing the tumor information density in the remaining data to be applied to the next step. After the 3-step removal, the 3D input is reduced to a minimum bounding box of the tumor region. It is used to finalize the prediction and then transformed into a 3D tumor mask, by means of gray level thresholding and low-pass-based morphological operations. The final prediction result is used to determine the critical threshold. The proposed system has been tested extensively with the data of more than one thousand patient cases in the datasets of BraTS 2018~21. The test results demonstrate that the predicted 2D histograms have a high degree of similarity with the true ones. The system delivers also very good tumor detection results, comparable to those of state-of-the-art CNN systems with mono-modality inputs, which is achieved at an extremely low computation cost and no need for training.
Coupling Global Context and Local Contents for Weakly-Supervised Semantic Segmentation
Apr 26, 2023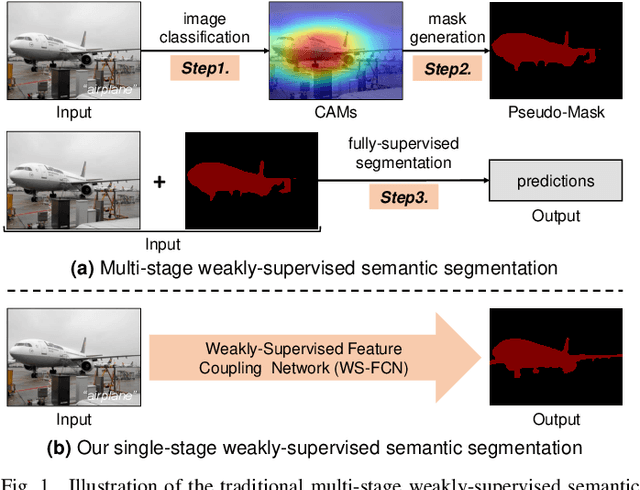

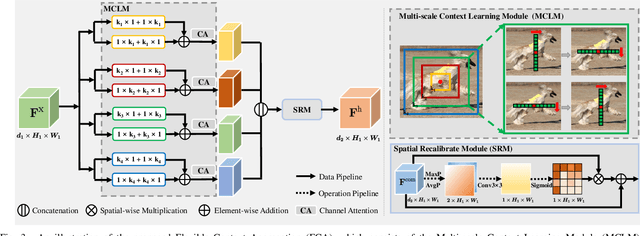
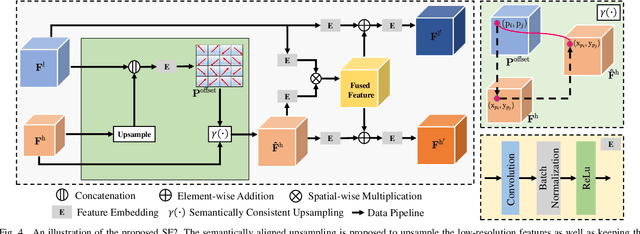
Abstract:Thanks to the advantages of the friendly annotations and the satisfactory performance, Weakly-Supervised Semantic Segmentation (WSSS) approaches have been extensively studied. Recently, the single-stage WSSS was awakened to alleviate problems of the expensive computational costs and the complicated training procedures in multi-stage WSSS. However, results of such an immature model suffer from problems of background incompleteness and object incompleteness. We empirically find that they are caused by the insufficiency of the global object context and the lack of the local regional contents, respectively. Under these observations, we propose a single-stage WSSS model with only the image-level class label supervisions, termed as Weakly Supervised Feature Coupling Network (WS-FCN), which can capture the multi-scale context formed from the adjacent feature grids, and encode the fine-grained spatial information from the low-level features into the high-level ones. Specifically, a flexible context aggregation module is proposed to capture the global object context in different granular spaces. Besides, a semantically consistent feature fusion module is proposed in a bottom-up parameter-learnable fashion to aggregate the fine-grained local contents. Based on these two modules, WS-FCN lies in a self-supervised end-to-end training fashion. Extensive experimental results on the challenging PASCAL VOC 2012 and MS COCO 2014 demonstrate the effectiveness and efficiency of WS-FCN, which can achieve state-of-the-art results by 65.02\% and 64.22\% mIoU on PASCAL VOC 2012 val set and test set, 34.12\% mIoU on MS COCO 2014 val set, respectively. The code and weight have been released at:https://github.com/ChunyanWang1/ws-fcn.
A Performance-Consistent and Computation-Efficient CNN System for High-Quality Automated Brain Tumor Segmentation
May 02, 2022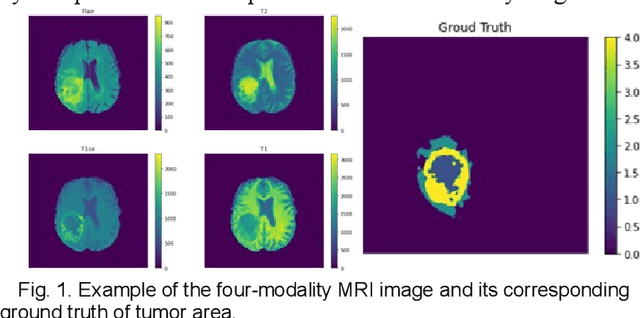
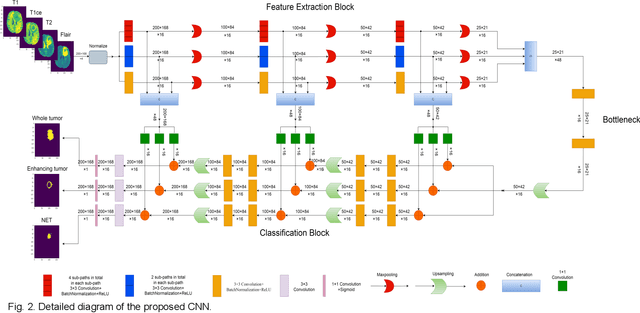
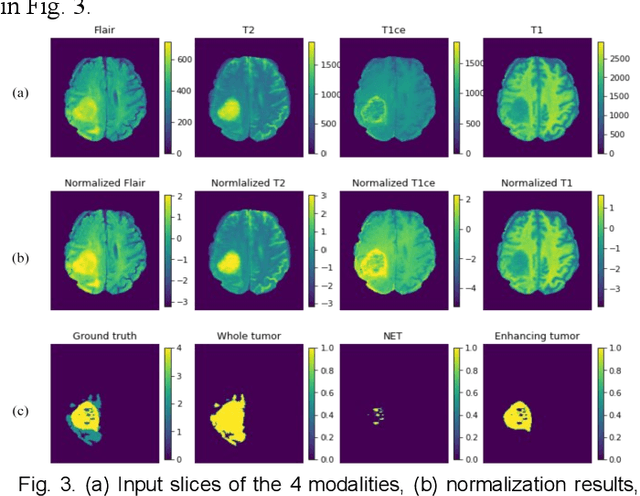
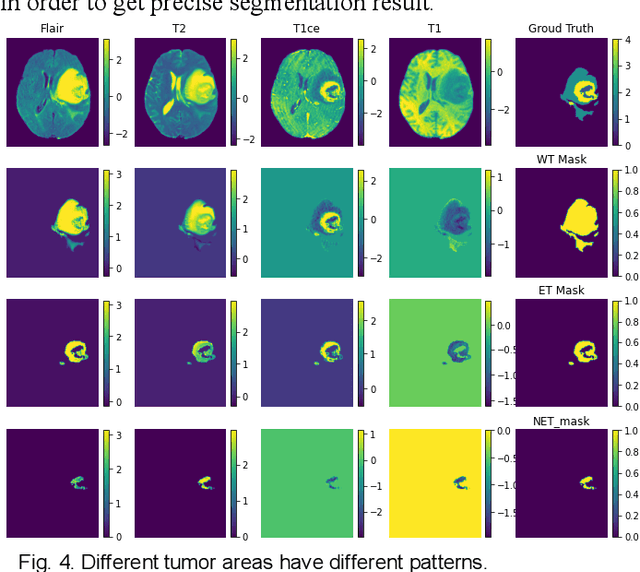
Abstract:The research on developing CNN-based fully-automated Brain-Tumor-Segmentation systems has been progressed rapidly. For the systems to be applicable in practice, a good The research on developing CNN-based fully-automated Brain-Tumor-Segmentation systems has been progressed rapidly. For the systems to be applicable in practice, a good processing quality and reliability are the must. Moreover, for wide applications of such systems, a minimization of computation complexity is desirable, which can also result in a minimization of randomness in computation and, consequently, a better performance consistency. To this end, the CNN in the proposed system has a unique structure with 2 distinguished characters. Firstly, the three paths of its feature extraction block are designed to extract, from the multi-modality input, comprehensive feature information of mono-modality, paired-modality and cross-modality data, respectively. Also, it has a particular three-branch classification block to identify the pixels of 4 classes. Each branch is trained separately so that the parameters are updated specifically with the corresponding ground truth data of a target tumor areas. The convolution layers of the system are custom-designed with specific purposes, resulting in a very simple config of 61,843 parameters in total. The proposed system is tested extensively with BraTS2018 and BraTS2019 datasets. The mean Dice scores, obtained from the ten experiments on BraTS2018 validation samples, are 0.787+0.003, 0.886+0.002, 0.801+0.007, for enhancing tumor, whole tumor and tumor core, respectively, and 0.751+0.007, 0.885+0.002, 0.776+0.004 on BraTS2019. The test results demonstrate that the proposed system is able to perform high-quality segmentation in a consistent manner. Furthermore, its extremely low computation complexity will facilitate its implementation/application in various environments.
A Computation-Efficient CNN System for High-Quality Brain Tumor Segmentation
Aug 07, 2020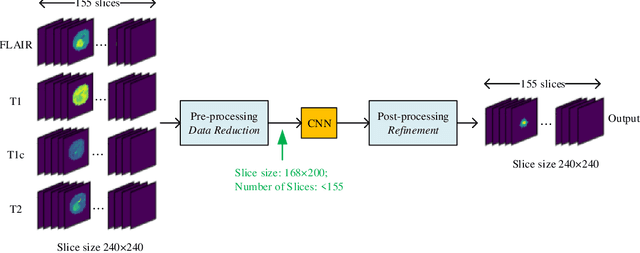
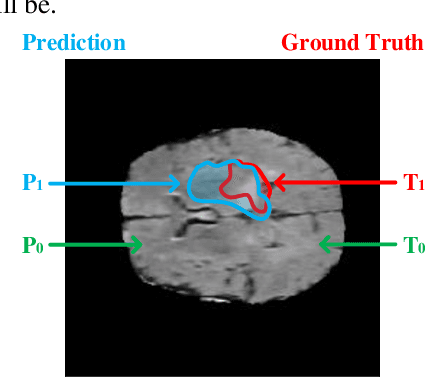
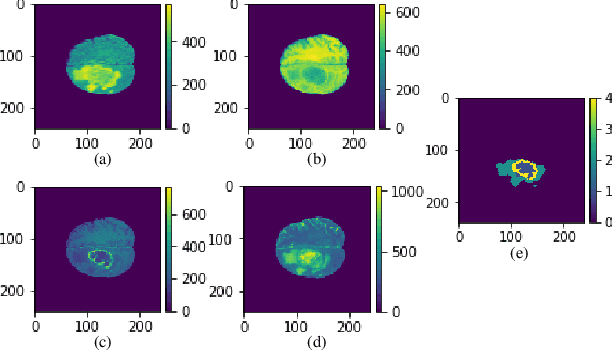
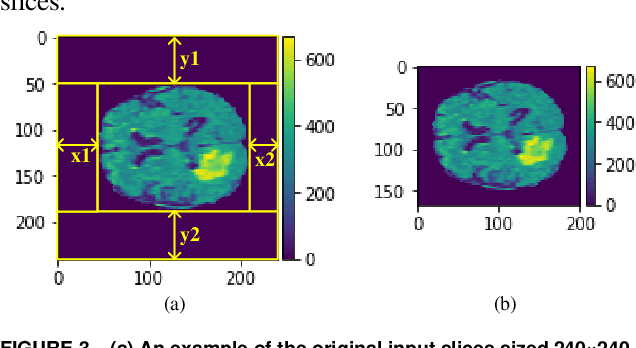
Abstract:In this paper, a Convolutional Neural Network (CNN) system is proposed for brain tumor segmentation. The system consists of three parts, a pre-processing block to reduce the data volume, an application-specific CNN(ASCNN) to segment tumor areas precisely, and a refinement block to detect/remove false positive pixels. The CNN, designed specifically for the task, has 7 convolution layers, 16 channels per layer, requiring only 11716 parameters. The convolutions combined with max-pooling in the first half of the CNN are performed to localize tumor areas. Two convolution modes, namely depthwise convolution and standard convolution, are performed in parallel in the first 2 layers to extract elementary features efficiently. For a fine classification of pixel-wise precision in the second half of the CNN, the feature maps are modulated by adding the individually weighted local feature maps generated in the first half of the CNN. The performance of the proposed system has been evaluated by an online platform with dataset of Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) 2018. Requiring a very low computation volume, the proposed system delivers a high segmentation quality indicated by its average Dice scores of 0.75, 0.88 and 0.76 for enhancing tumor, whole tumor and tumor core, respectively, and also by the median Dice scores of 0.85, 0.92, and 0.86. The consistency in system performance has also been measured, demonstrating that the system is able to reproduce almost the same output to the same input after retraining. The simple structure of the proposed system facilitates its implementation in computation restricted environment, and a wide range of applications can thus be expected.
SdcNet: A Computation-Efficient CNN for Object Recognition
May 05, 2018

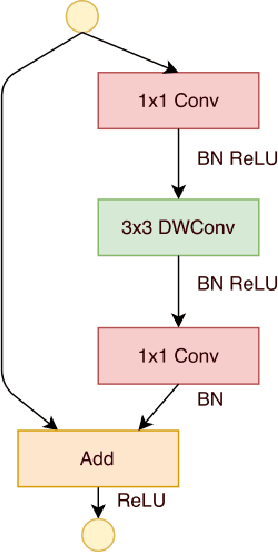
Abstract:Extracting features from a huge amount of data for object recognition is a challenging task. Convolution neural network can be used to meet the challenge, but it often requires a large number of computation resources. In this paper, a computation-efficient convolutional module, named SdcBlock, is proposed and based on it, the convolution network SdcNet is introduced for object recognition tasks. In the proposed module, optimized successive depthwise convolutions supported by appropriate data management is applied in order to generate vectors containing high density and more varieties of feature information. The hyperparameters can be easily adjusted to suit varieties of tasks under different computation restrictions without significantly jeopardizing the performance. The experiments have shown that SdcNet achieved an error rate of 5.60% in CIFAR-10 with only 55M Flops and also reduced further the error rate to 5.24% using a moderate volume of 103M Flops. The expected computation efficiency of the SdcNet has been confirmed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge