Yalun Hu
Adversarial Attacks on Spiking Convolutional Networks for Event-based Vision
Oct 06, 2021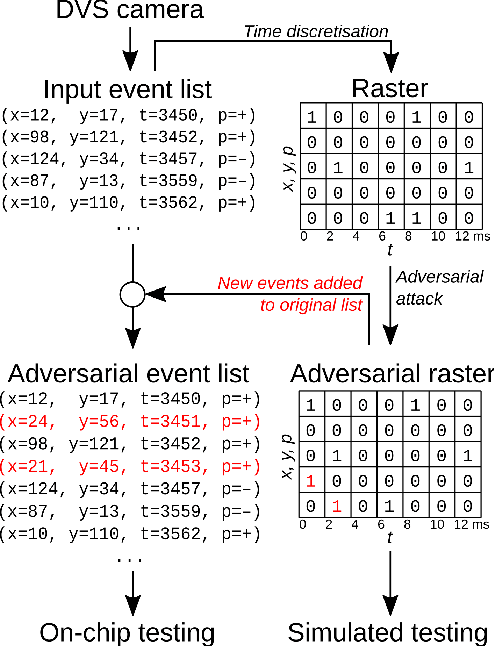
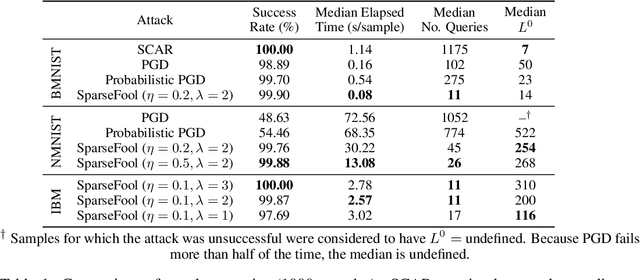
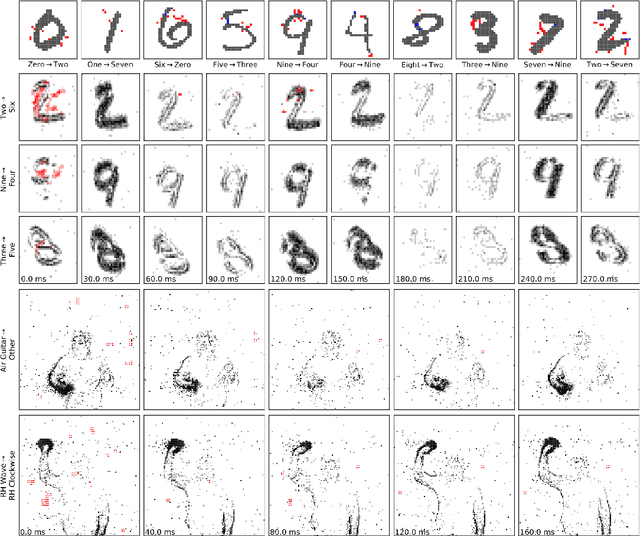
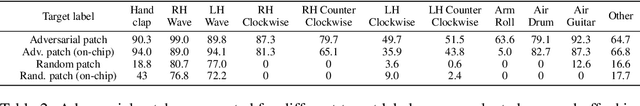
Abstract:Event-based sensing using dynamic vision sensors is gaining traction in low-power vision applications. Spiking neural networks work well with the sparse nature of event-based data and suit deployment on low-power neuromorphic hardware. Being a nascent field, the sensitivity of spiking neural networks to potentially malicious adversarial attacks has received very little attention so far. In this work, we show how white-box adversarial attack algorithms can be adapted to the discrete and sparse nature of event-based visual data, and to the continuous-time setting of spiking neural networks. We test our methods on the N-MNIST and IBM Gestures neuromorphic vision datasets and show adversarial perturbations achieve a high success rate, by injecting a relatively small number of appropriately placed events. We also verify, for the first time, the effectiveness of these perturbations directly on neuromorphic hardware. Finally, we discuss the properties of the resulting perturbations and possible future directions.
A Novel Application of Image-to-Image Translation: Chromosome Straightening Framework by Learning from a Single Image
Mar 04, 2021



Abstract:In medical imaging, chromosome straightening plays a significant role in the pathological study of chromosomes and in the development of cytogenetic maps. Whereas different approaches exist for the straightening task, they are mostly geometric algorithms whose outputs are characterized by jagged edges or fragments with discontinued banding patterns. To address the flaws in the geometric algorithms, we propose a novel framework based on image-to-image translation to learn a pertinent mapping dependence for synthesizing straightened chromosomes with uninterrupted banding patterns and preserved details. In addition, to avoid the pitfall of deficient input chromosomes, we construct an augmented dataset using only one single curved chromosome image for training models. Based on this framework, we apply two popular image-to-image translation architectures, U-shape networks and conditional generative adversarial networks, to assess its efficacy. Experiments on a dataset comprising of 642 real-world chromosomes demonstrate the superiority of our framework as compared to the geometric method in straightening performance by rendering realistic and continued chromosome details. Furthermore, our straightened results improve the chromosome classification, achieving 0.98%-1.39% in mean accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge