Wing Yee Au
Harnessing Rich Multi-Modal Data for Spatial-Temporal Homophily-Embedded Graph Learning Across Domains and Localities
Dec 11, 2025Abstract:Modern cities are increasingly reliant on data-driven insights to support decision making in areas such as transportation, public safety and environmental impact. However, city-level data often exists in heterogeneous formats, collected independently by local agencies with diverse objectives and standards. Despite their numerous, wide-ranging, and uniformly consumable nature, national-level datasets exhibit significant heterogeneity and multi-modality. This research proposes a heterogeneous data pipeline that performs cross-domain data fusion over time-varying, spatial-varying and spatial-varying time-series datasets. We aim to address complex urban problems across multiple domains and localities by harnessing the rich information over 50 data sources. Specifically, our data-learning module integrates homophily from spatial-varying dataset into graph-learning, embedding information of various localities into models. We demonstrate the generalizability and flexibility of the framework through five real-world observations using a variety of publicly accessible datasets (e.g., ride-share, traffic crash, and crime reports) collected from multiple cities. The results show that our proposed framework demonstrates strong predictive performance while requiring minimal reconfiguration when transferred to new localities or domains. This research advances the goal of building data-informed urban systems in a scalable way, addressing one of the most pressing challenges in smart city analytics.
Personalized Real-time Jargon Support for Online Meetings
Aug 13, 2025



Abstract:Effective interdisciplinary communication is frequently hindered by domain-specific jargon. To explore the jargon barriers in-depth, we conducted a formative diary study with 16 professionals, revealing critical limitations in current jargon-management strategies during workplace meetings. Based on these insights, we designed ParseJargon, an interactive LLM-powered system providing real-time personalized jargon identification and explanations tailored to users' individual backgrounds. A controlled experiment comparing ParseJargon against baseline (no support) and general-purpose (non-personalized) conditions demonstrated that personalized jargon support significantly enhanced participants' comprehension, engagement, and appreciation of colleagues' work, whereas general-purpose support negatively affected engagement. A follow-up field study validated ParseJargon's usability and practical value in real-time meetings, highlighting both opportunities and limitations for real-world deployment. Our findings contribute insights into designing personalized jargon support tools, with implications for broader interdisciplinary and educational applications.
A Latent Diffusion Model for Protein Structure Generation
May 06, 2023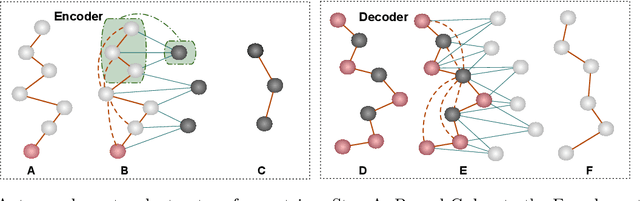
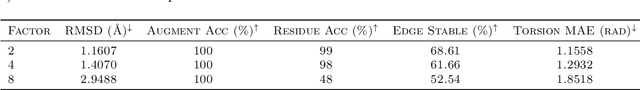
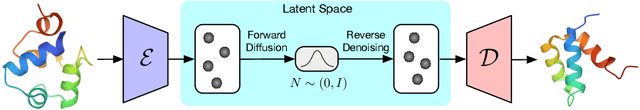
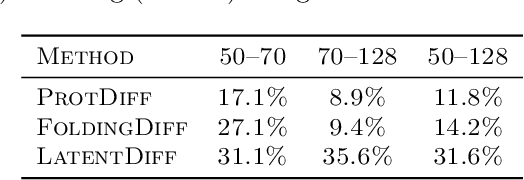
Abstract:Proteins are complex biomolecules that perform a variety of crucial functions within living organisms. Designing and generating novel proteins can pave the way for many future synthetic biology applications, including drug discovery. However, it remains a challenging computational task due to the large modeling space of protein structures. In this study, we propose a latent diffusion model that can reduce the complexity of protein modeling while flexibly capturing the distribution of natural protein structures in a condensed latent space. Specifically, we propose an equivariant protein autoencoder that embeds proteins into a latent space and then uses an equivariant diffusion model to learn the distribution of the latent protein representations. Experimental results demonstrate that our method can effectively generate novel protein backbone structures with high designability and efficiency.
Automated Data Augmentations for Graph Classification
Mar 19, 2022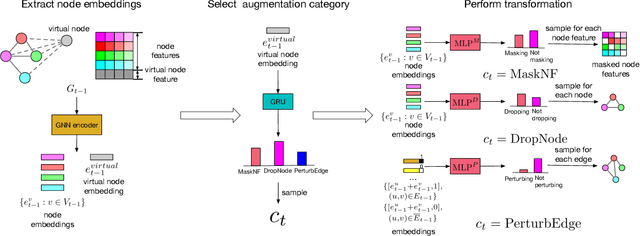
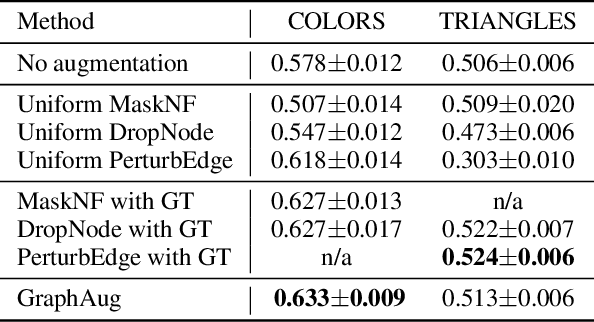
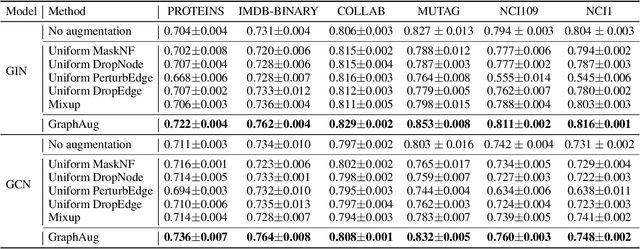
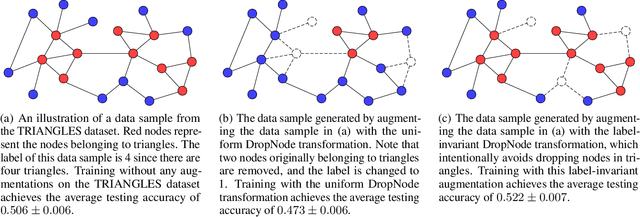
Abstract:Data augmentations are effective in improving the invariance of learning machines. We argue that the corechallenge of data augmentations lies in designing data transformations that preserve labels. This is relativelystraightforward for images, but much more challenging for graphs. In this work, we propose GraphAug, a novelautomated data augmentation method aiming at computing label-invariant augmentations for graph classification.Instead of using uniform transformations as in existing studies, GraphAug uses an automated augmentationmodel to avoid compromising critical label-related information of the graph, thereby producing label-invariantaugmentations at most times. To ensure label-invariance, we develop a training method based on reinforcementlearning to maximize an estimated label-invariance probability. Comprehensive experiments show that GraphAugoutperforms previous graph augmentation methods on various graph classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge