Kanji Uchino
A Latent Diffusion Model for Protein Structure Generation
May 06, 2023
Abstract:Proteins are complex biomolecules that perform a variety of crucial functions within living organisms. Designing and generating novel proteins can pave the way for many future synthetic biology applications, including drug discovery. However, it remains a challenging computational task due to the large modeling space of protein structures. In this study, we propose a latent diffusion model that can reduce the complexity of protein modeling while flexibly capturing the distribution of natural protein structures in a condensed latent space. Specifically, we propose an equivariant protein autoencoder that embeds proteins into a latent space and then uses an equivariant diffusion model to learn the distribution of the latent protein representations. Experimental results demonstrate that our method can effectively generate novel protein backbone structures with high designability and efficiency.
Generating 3D Molecules for Target Protein Binding
Apr 19, 2022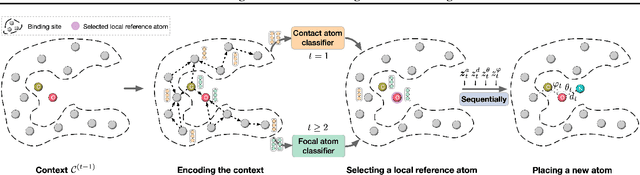
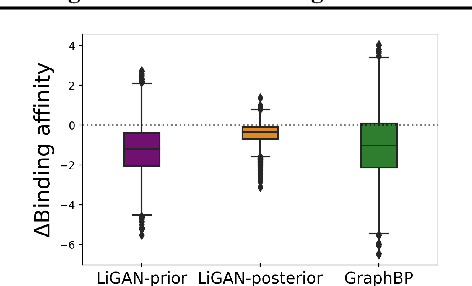
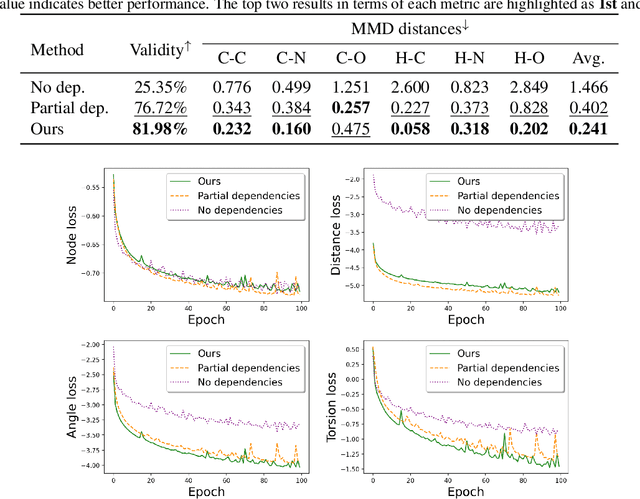
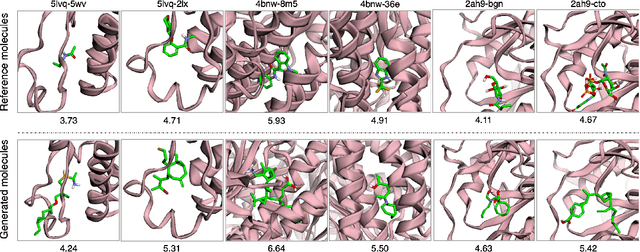
Abstract:A fundamental problem in drug discovery is to design molecules that bind to specific proteins. To tackle this problem using machine learning methods, here we propose a novel and effective framework, known as GraphBP, to generate 3D molecules that bind to given proteins by placing atoms of specific types and locations to the given binding site one by one. In particular, at each step, we first employ a 3D graph neural network to obtain geometry-aware and chemically informative representations from the intermediate contextual information. Such context includes the given binding site and atoms placed in the previous steps. Second, to preserve the desirable equivariance property, we select a local reference atom according to the designed auxiliary classifiers and then construct a local spherical coordinate system. Finally, to place a new atom, we generate its atom type and relative location w.r.t. the constructed local coordinate system via a flow model. We also consider generating the variables of interest sequentially to capture the underlying dependencies among them. Experiments demonstrate that our GraphBP is effective to generate 3D molecules with binding ability to target protein binding sites. Our implementation is available at https://github.com/divelab/GraphBP.
Automated Data Augmentations for Graph Classification
Mar 19, 2022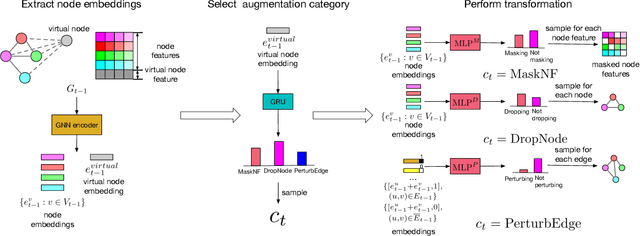
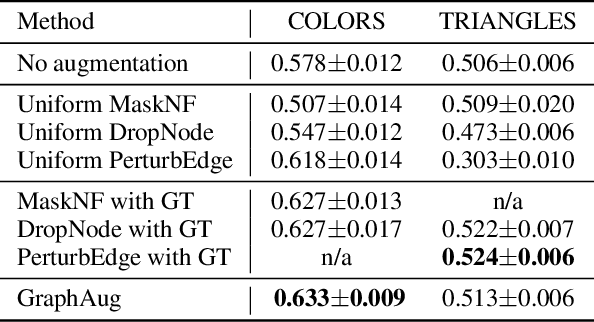
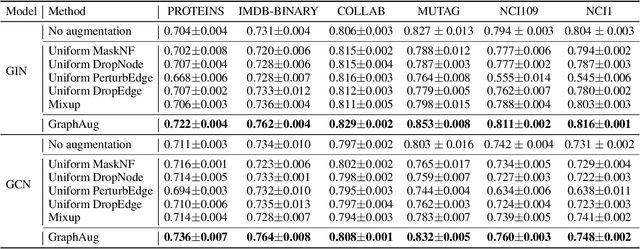
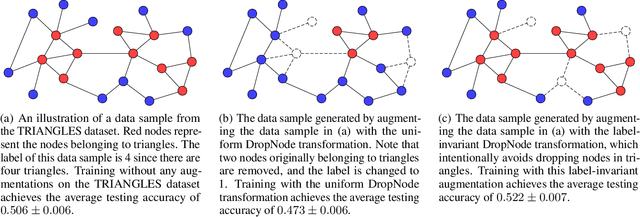
Abstract:Data augmentations are effective in improving the invariance of learning machines. We argue that the corechallenge of data augmentations lies in designing data transformations that preserve labels. This is relativelystraightforward for images, but much more challenging for graphs. In this work, we propose GraphAug, a novelautomated data augmentation method aiming at computing label-invariant augmentations for graph classification.Instead of using uniform transformations as in existing studies, GraphAug uses an automated augmentationmodel to avoid compromising critical label-related information of the graph, thereby producing label-invariantaugmentations at most times. To ensure label-invariance, we develop a training method based on reinforcementlearning to maximize an estimated label-invariance probability. Comprehensive experiments show that GraphAugoutperforms previous graph augmentation methods on various graph classification tasks.
Biases in Generative Art---A Causal Look from the Lens of Art History
Oct 26, 2020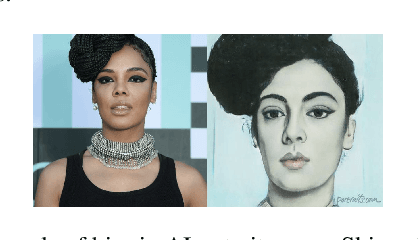


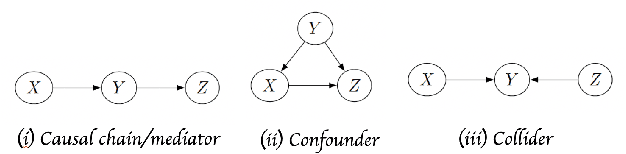
Abstract:With rapid progress in artificial intelligence (AI), popularity of generative art has grown substantially. From creating paintings to generating novel art styles, AI based generative art has showcased a variety of applications. However, there has been little focus concerning the ethical impacts of AI based generative art. In this work, we investigate biases in the generative art AI pipeline right from those that can originate due to improper problem formulation to those related to algorithm design. Viewing from the lens of art history, we discuss the socio-cultural impacts of these biases. Leveraging causal models, we highlight how current methods fall short in modeling the process of art creation and thus contribute to various types of biases. We illustrate the same through case studies. To the best of our knowledge, this is the first extensive analysis that investigates biases in the generative art AI pipeline from the perspective of art history. We hope our work sparks interdisciplinary discussions related to accountability of generative art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge