Walt Williams
A General-Purpose Self-Supervised Model for Computational Pathology
Aug 29, 2023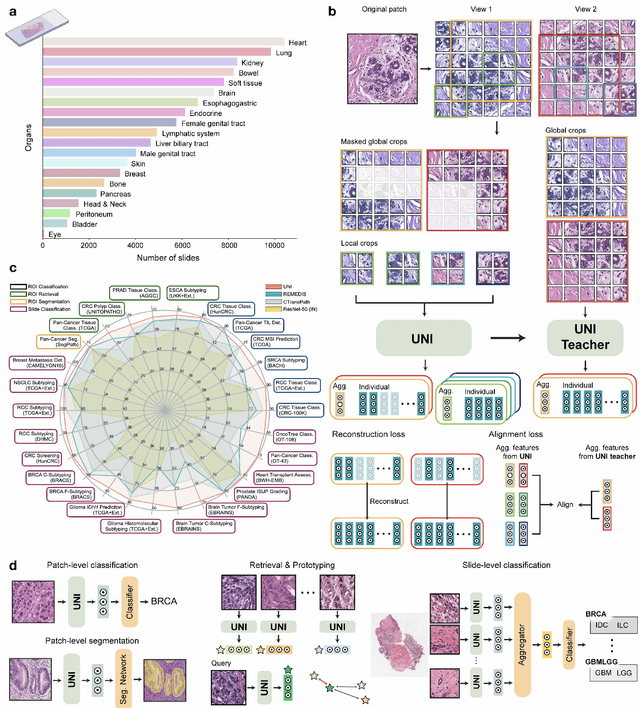

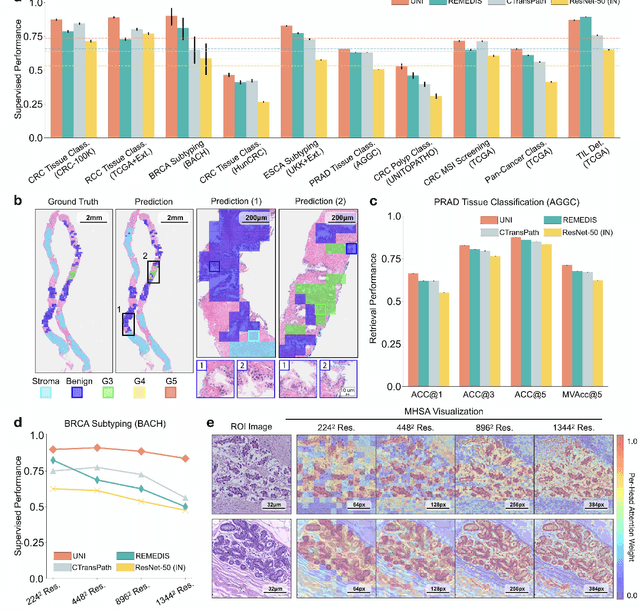

Abstract:Tissue phenotyping is a fundamental computational pathology (CPath) task in learning objective characterizations of histopathologic biomarkers in anatomic pathology. However, whole-slide imaging (WSI) poses a complex computer vision problem in which the large-scale image resolutions of WSIs and the enormous diversity of morphological phenotypes preclude large-scale data annotation. Current efforts have proposed using pretrained image encoders with either transfer learning from natural image datasets or self-supervised pretraining on publicly-available histopathology datasets, but have not been extensively developed and evaluated across diverse tissue types at scale. We introduce UNI, a general-purpose self-supervised model for pathology, pretrained using over 100 million tissue patches from over 100,000 diagnostic haematoxylin and eosin-stained WSIs across 20 major tissue types, and evaluated on 33 representative CPath clinical tasks in CPath of varying diagnostic difficulties. In addition to outperforming previous state-of-the-art models, we demonstrate new modeling capabilities in CPath such as resolution-agnostic tissue classification, slide classification using few-shot class prototypes, and disease subtyping generalization in classifying up to 108 cancer types in the OncoTree code classification system. UNI advances unsupervised representation learning at scale in CPath in terms of both pretraining data and downstream evaluation, enabling data-efficient AI models that can generalize and transfer to a gamut of diagnostically-challenging tasks and clinical workflows in anatomic pathology.
Predicting Ejection Fraction from Chest X-rays Using Computer Vision for Diagnosing Heart Failure
Dec 19, 2022



Abstract:Heart failure remains a major public health challenge with growing costs. Ejection fraction (EF) is a key metric for the diagnosis and management of heart failure however estimation of EF using echocardiography remains expensive for the healthcare system and subject to intra/inter operator variability. While chest x-rays (CXR) are quick, inexpensive, and require less expertise, they do not provide sufficient information to the human eye to estimate EF. This work explores the efficacy of computer vision techniques to predict reduced EF solely from CXRs. We studied a dataset of 3488 CXRs from the MIMIC CXR-jpg (MCR) dataset. Our work establishes benchmarks using multiple state-of-the-art convolutional neural network architectures. The subsequent analysis shows increasing model sizes from 8M to 23M parameters improved classification performance without overfitting the dataset. We further show how data augmentation techniques such as CXR rotation and random cropping further improves model performance another ~5%. Finally, we conduct an error analysis using saliency maps and Grad-CAMs to better understand the failure modes of convolutional models on this task.
AMDet: A Tool for Mitotic Cell Detection in Histopathology Slides
Aug 08, 2021
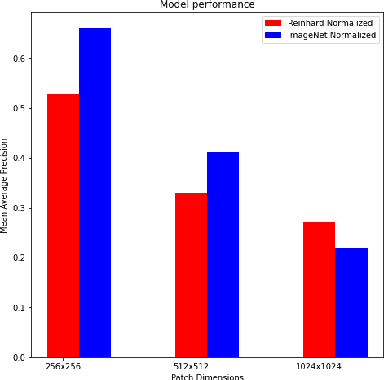
Abstract:Breast Cancer is the most prevalent cancer in the world. The World Health Organization reports that the disease still affects a significant portion of the developing world citing increased mortality rates in the majority of low to middle income countries. The most popular protocol pathologists use for diagnosing breast cancer is the Nottingham grading system which grades the proliferation of tumors based on 3 major criteria, the most important of them being mitotic cell count. The way in which pathologists evaluate mitotic cell count is to subjectively and qualitatively analyze cells present in stained slides of tissue and make a decision on its mitotic state i.e. is it mitotic or not?This process is extremely inefficient and tiring for pathologists and so an efficient, accurate, and fully automated tool to aid with the diagnosis is extremely desirable. Fortunately, creating such a tool is made significantly easier with the AutoML tool available from Microsoft Azure, however to the best of our knowledge the AutoML tool has never been formally evaluated for use in mitotic cell detection in histopathology images. This paper serves as an evaluation of the AutoML tool for this purpose and will provide a first look on how the tool handles this challenging problem. All code is available athttps://github.com/WaltAFWilliams/AMDet
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge