Vivek Vaidya
How Good LLM-Generated Password Policies Are?
Jun 10, 2025



Abstract:Generative AI technologies, particularly Large Language Models (LLMs), are rapidly being adopted across industry, academia, and government sectors, owing to their remarkable capabilities in natural language processing. However, despite their strengths, the inconsistency and unpredictability of LLM outputs present substantial challenges, especially in security-critical domains such as access control. One critical issue that emerges prominently is the consistency of LLM-generated responses, which is paramount for ensuring secure and reliable operations. In this paper, we study the application of LLMs within the context of Cybersecurity Access Control Systems. Specifically, we investigate the consistency and accuracy of LLM-generated password policies, translating natural language prompts into executable pwquality.conf configuration files. Our experimental methodology adopts two distinct approaches: firstly, we utilize pre-trained LLMs to generate configuration files purely from natural language prompts without additional guidance. Secondly, we provide these models with official pwquality.conf documentation to serve as an informative baseline. We systematically assess the soundness, accuracy, and consistency of these AI-generated configurations. Our findings underscore significant challenges in the current generation of LLMs and contribute valuable insights into refining the deployment of LLMs in Access Control Systems.
Automated Consistency Analysis of LLMs
Feb 10, 2025



Abstract:Generative AI (Gen AI) with large language models (LLMs) are being widely adopted across the industry, academia and government. Cybersecurity is one of the key sectors where LLMs can be and/or are already being used. There are a number of problems that inhibit the adoption of trustworthy Gen AI and LLMs in cybersecurity and such other critical areas. One of the key challenge to the trustworthiness and reliability of LLMs is: how consistent an LLM is in its responses? In this paper, we have analyzed and developed a formal definition of consistency of responses of LLMs. We have formally defined what is consistency of responses and then develop a framework for consistency evaluation. The paper proposes two approaches to validate consistency: self-validation, and validation across multiple LLMs. We have carried out extensive experiments for several LLMs such as GPT4oMini, GPT3.5, Gemini, Cohere, and Llama3, on a security benchmark consisting of several cybersecurity questions: informational and situational. Our experiments corroborate the fact that even though these LLMs are being considered and/or already being used for several cybersecurity tasks today, they are often inconsistent in their responses, and thus are untrustworthy and unreliable for cybersecurity.
* 10 pages, 12 figures, 3 tables, 3 algorithms
Understanding the Mechanisms of Deep Transfer Learning for Medical Images
Apr 20, 2017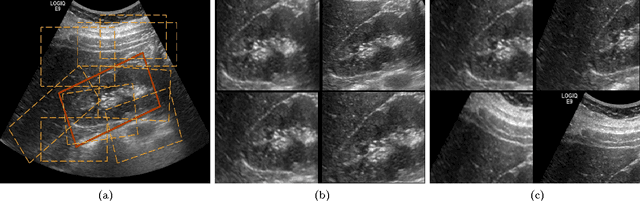
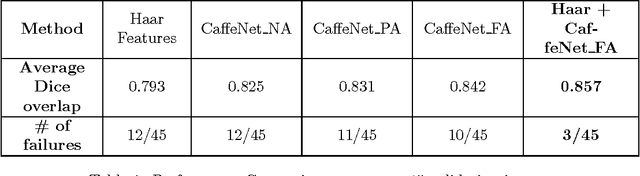
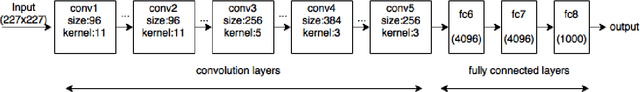

Abstract:The ability to automatically learn task specific feature representations has led to a huge success of deep learning methods. When large training data is scarce, such as in medical imaging problems, transfer learning has been very effective. In this paper, we systematically investigate the process of transferring a Convolutional Neural Network, trained on ImageNet images to perform image classification, to kidney detection problem in ultrasound images. We study how the detection performance depends on the extent of transfer. We show that a transferred and tuned CNN can outperform a state-of-the-art feature engineered pipeline and a hybridization of these two techniques achieves 20\% higher performance. We also investigate how the evolution of intermediate response images from our network. Finally, we compare these responses to state-of-the-art image processing filters in order to gain greater insight into how transfer learning is able to effectively manage widely varying imaging regimes.
Filter sharing: Efficient learning of parameters for volumetric convolutions
Dec 08, 2016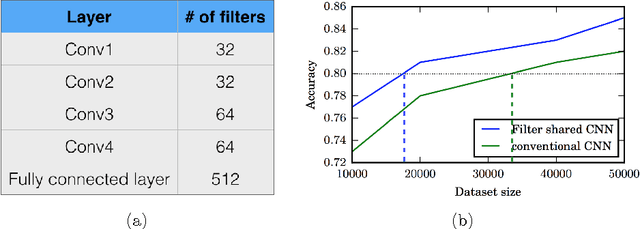

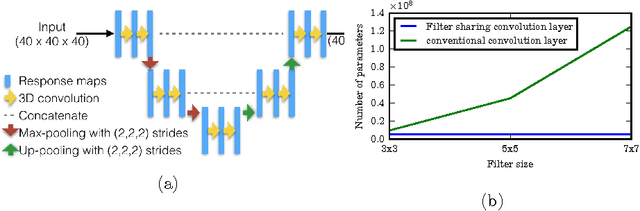
Abstract:Typical convolutional neural networks (CNNs) have several millions of parameters and require a large amount of annotated data to train them. In medical applications where training data is hard to come by, these sophisticated machine learning models are difficult to train. In this paper, we propose a method to reduce the inherent complexity of CNNs during training by exploiting the significant redundancy that is noticed in the learnt CNN filters. Our method relies on finding a small set of filters and mixing coefficients to derive every filter in each convolutional layer at the time of training itself, thereby reducing the number of parameters to be trained. We consider the problem of 3D lung nodule segmentation in CT images and demonstrate the effectiveness of our method in achieving good results with only few training examples.
An SVM Based Approach for Cardiac View Planning
Jul 11, 2014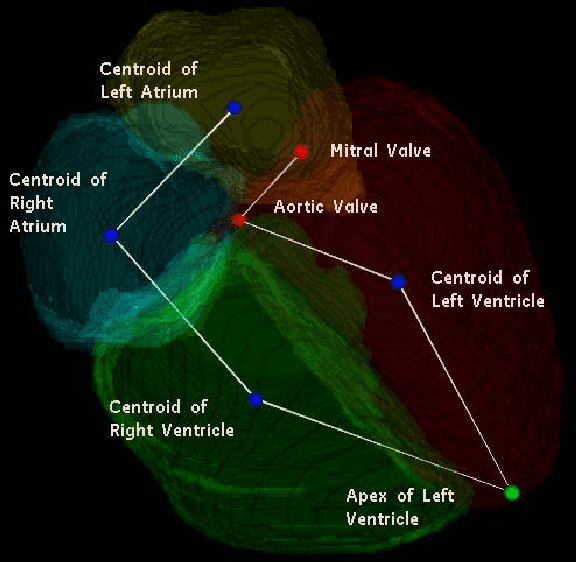

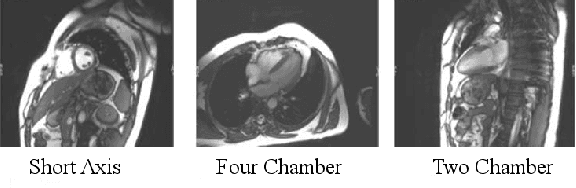

Abstract:We consider the problem of automatically prescribing oblique planes (short axis, 4 chamber and 2 chamber views) in Cardiac Magnetic Resonance Imaging (MRI). A concern with technologist-driven acquisitions of these planes is the quality and time taken for the total examination. We propose an automated solution incorporating anatomical features external to the cardiac region. The solution uses support vector machine regression models wherein complexity and feature selection are optimized using multi-objective genetic algorithms. Additionally, we examine the robustness of our approach by training our models on images with additive Rician-Gaussian mixtures at varying Signal to Noise (SNR) levels. Our approach has shown promising results, with an angular deviation of less than 15 degrees on 90% cases across oblique planes, measured in terms of average 6-fold cross validation performance -- this is generally within acceptable bounds of variation as specified by clinicians.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge