Sheshadri Thiruvenkadam
Understanding the Mechanisms of Deep Transfer Learning for Medical Images
Apr 20, 2017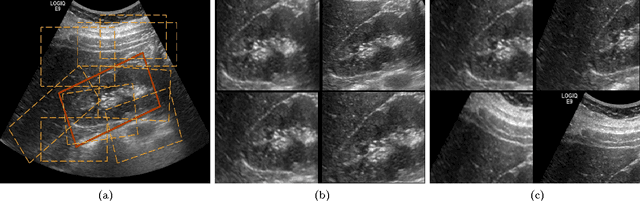
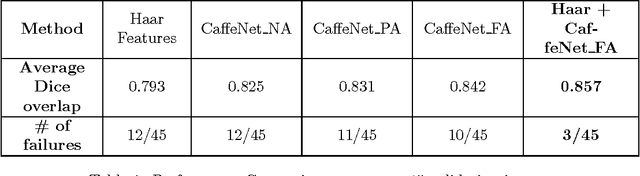
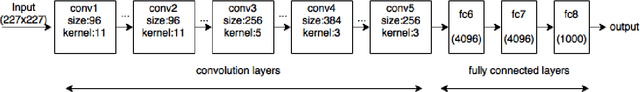

Abstract:The ability to automatically learn task specific feature representations has led to a huge success of deep learning methods. When large training data is scarce, such as in medical imaging problems, transfer learning has been very effective. In this paper, we systematically investigate the process of transferring a Convolutional Neural Network, trained on ImageNet images to perform image classification, to kidney detection problem in ultrasound images. We study how the detection performance depends on the extent of transfer. We show that a transferred and tuned CNN can outperform a state-of-the-art feature engineered pipeline and a hybridization of these two techniques achieves 20\% higher performance. We also investigate how the evolution of intermediate response images from our network. Finally, we compare these responses to state-of-the-art image processing filters in order to gain greater insight into how transfer learning is able to effectively manage widely varying imaging regimes.
Filter sharing: Efficient learning of parameters for volumetric convolutions
Dec 08, 2016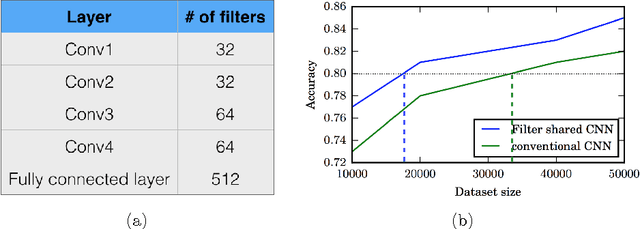

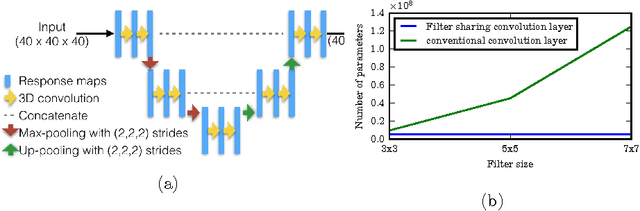
Abstract:Typical convolutional neural networks (CNNs) have several millions of parameters and require a large amount of annotated data to train them. In medical applications where training data is hard to come by, these sophisticated machine learning models are difficult to train. In this paper, we propose a method to reduce the inherent complexity of CNNs during training by exploiting the significant redundancy that is noticed in the learnt CNN filters. Our method relies on finding a small set of filters and mixing coefficients to derive every filter in each convolutional layer at the time of training itself, thereby reducing the number of parameters to be trained. We consider the problem of 3D lung nodule segmentation in CT images and demonstrate the effectiveness of our method in achieving good results with only few training examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge