Rahul Venkataramani
Synthetic Simplicity: Unveiling Bias in Medical Data Augmentation
Jul 31, 2024Abstract:Synthetic data is becoming increasingly integral in data-scarce fields such as medical imaging, serving as a substitute for real data. However, its inherent statistical characteristics can significantly impact downstream tasks, potentially compromising deployment performance. In this study, we empirically investigate this issue and uncover a critical phenomenon: downstream neural networks often exploit spurious distinctions between real and synthetic data when there is a strong correlation between the data source and the task label. This exploitation manifests as \textit{simplicity bias}, where models overly rely on superficial features rather than genuine task-related complexities. Through principled experiments, we demonstrate that the source of data (real vs.\ synthetic) can introduce spurious correlating factors leading to poor performance during deployment when the correlation is absent. We first demonstrate this vulnerability on a digit classification task, where the model spuriously utilizes the source of data instead of the digit to provide an inference. We provide further evidence of this phenomenon in a medical imaging problem related to cardiac view classification in echocardiograms, particularly distinguishing between 2-chamber and 4-chamber views. Given the increasing role of utilizing synthetic datasets, we hope that our experiments serve as effective guidelines for the utilization of synthetic datasets in model training.
Task-driven Prompt Evolution for Foundation Models
Oct 26, 2023Abstract:Promptable foundation models, particularly Segment Anything Model (SAM), have emerged as a promising alternative to the traditional task-specific supervised learning for image segmentation. However, many evaluation studies have found that their performance on medical imaging modalities to be underwhelming compared to conventional deep learning methods. In the world of large pre-trained language and vision-language models, learning prompt from downstream tasks has achieved considerable success in improving performance. In this work, we propose a plug-and-play Prompt Optimization Technique for foundation models like SAM (SAMPOT) that utilizes the downstream segmentation task to optimize the human-provided prompt to obtain improved performance. We demonstrate the utility of SAMPOT on lung segmentation in chest X-ray images and obtain an improvement on a significant number of cases ($\sim75\%$) over human-provided initial prompts. We hope this work will lead to further investigations in the nascent field of automatic visual prompt-tuning.
Towards Continuous Domain adaptation for Healthcare
Dec 04, 2018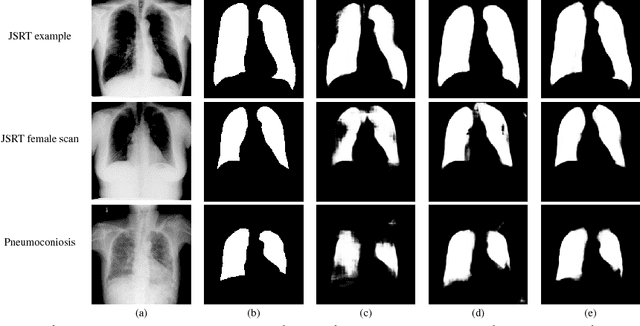
Abstract:Deep learning algorithms have demonstrated tremendous success on challenging medical imaging problems. However, post-deployment, these algorithms are susceptible to data distribution variations owing to \emph{limited data issues} and \emph{diversity} in medical images. In this paper, we propose \emph{ContextNets}, a generic memory-augmented neural network framework for semantic segmentation to achieve continuous domain adaptation without the necessity of retraining. Unlike existing methods which require access to entire source and target domain images, our algorithm can adapt to a target domain with a few similar images. We condition the inference on any new input with features computed on its support set of images (and masks, if available) through contextual embeddings to achieve site-specific adaptation. We demonstrate state-of-the-art domain adaptation performance on the X-ray lung segmentation problem from three independent cohorts that differ in disease type, gender, contrast and intensity variations.
Understanding the Mechanisms of Deep Transfer Learning for Medical Images
Apr 20, 2017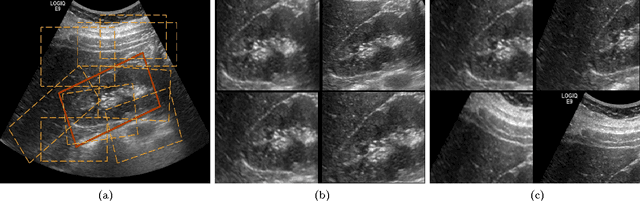
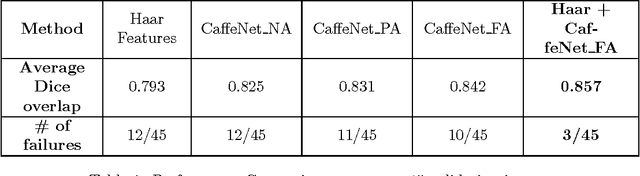
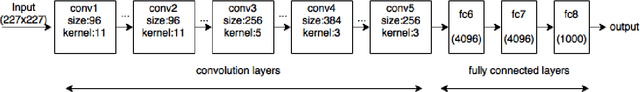

Abstract:The ability to automatically learn task specific feature representations has led to a huge success of deep learning methods. When large training data is scarce, such as in medical imaging problems, transfer learning has been very effective. In this paper, we systematically investigate the process of transferring a Convolutional Neural Network, trained on ImageNet images to perform image classification, to kidney detection problem in ultrasound images. We study how the detection performance depends on the extent of transfer. We show that a transferred and tuned CNN can outperform a state-of-the-art feature engineered pipeline and a hybridization of these two techniques achieves 20\% higher performance. We also investigate how the evolution of intermediate response images from our network. Finally, we compare these responses to state-of-the-art image processing filters in order to gain greater insight into how transfer learning is able to effectively manage widely varying imaging regimes.
Filter sharing: Efficient learning of parameters for volumetric convolutions
Dec 08, 2016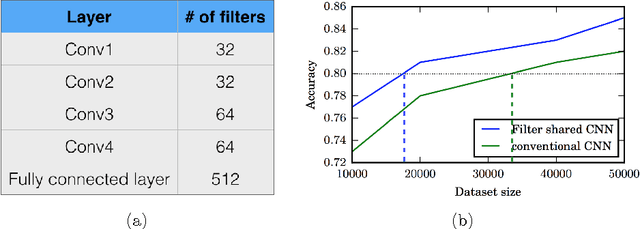

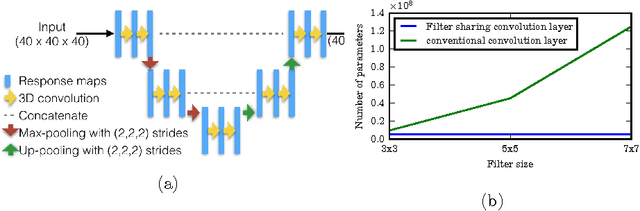
Abstract:Typical convolutional neural networks (CNNs) have several millions of parameters and require a large amount of annotated data to train them. In medical applications where training data is hard to come by, these sophisticated machine learning models are difficult to train. In this paper, we propose a method to reduce the inherent complexity of CNNs during training by exploiting the significant redundancy that is noticed in the learnt CNN filters. Our method relies on finding a small set of filters and mixing coefficients to derive every filter in each convolutional layer at the time of training itself, thereby reducing the number of parameters to be trained. We consider the problem of 3D lung nodule segmentation in CT images and demonstrate the effectiveness of our method in achieving good results with only few training examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge