Vijay Raghavan
Beyond the Hype: Embeddings vs. Prompting for Multiclass Classification Tasks
Apr 09, 2025Abstract:Are traditional classification approaches irrelevant in this era of AI hype? We show that there are multiclass classification problems where predictive models holistically outperform LLM prompt-based frameworks. Given text and images from home-service project descriptions provided by Thumbtack customers, we build embeddings-based softmax models that predict the professional category (e.g., handyman, bathroom remodeling) associated with each problem description. We then compare against prompts that ask state-of-the-art LLM models to solve the same problem. We find that the embeddings approach outperforms the best LLM prompts in terms of accuracy, calibration, latency, and financial cost. In particular, the embeddings approach has 49.5% higher accuracy than the prompting approach, and its superiority is consistent across text-only, image-only, and text-image problem descriptions. Furthermore, it yields well-calibrated probabilities, which we later use as confidence signals to provide contextualized user experience during deployment. On the contrary, prompting scores are overly uninformative. Finally, the embeddings approach is 14 and 81 times faster than prompting in processing images and text respectively, while under realistic deployment assumptions, it can be up to 10 times cheaper. Based on these results, we deployed a variation of the embeddings approach, and through A/B testing we observed performance consistent with our offline analysis. Our study shows that for multiclass classification problems that can leverage proprietary datasets, an embeddings-based approach may yield unequivocally better results. Hence, scientists, practitioners, engineers, and business leaders can use our study to go beyond the hype and consider appropriate predictive models for their classification use cases.
Integrating Secondary Structures Information into Triangular Spatial Relationships (TSR) for Advanced Protein Classification
Nov 19, 2024



Abstract:Protein structures represent the key to deciphering biological functions. The more detailed form of similarity among these proteins is sometimes overlooked by the conventional structural comparison methods. In contrast, further advanced methods, such as Triangular Spatial Relationship (TSR), have been demonstrated to make finer differentiations. Still, the classical implementation of TSR does not provide for the integration of secondary structure information, which is important for a more detailed understanding of the folding pattern of a protein. To overcome these limitations, we developed the SSE-TSR approach. The proposed method integrates secondary structure elements (SSEs) into TSR-based protein representations. This allows an enriched representation of protein structures by considering 18 different combinations of helix, strand, and coil arrangements. Our results show that using SSEs improves the accuracy and reliability of protein classification to varying degrees. We worked with two large protein datasets of 9.2K and 7.8K samples, respectively. We applied the SSE-TSR approach and used a neural network model for classification. Interestingly, introducing SSEs improved performance statistics for Dataset 1, with accuracy moving from 96.0% to 98.3%. For Dataset 2, where the performance statistics were already good, further small improvements were found with the introduction of SSE, giving an accuracy of 99.5% compared to 99.4%. These results show that SSE integration can dramatically improve TSR key discrimination, with significant benefits in datasets with low initial accuracies and only incremental gains in those with high baseline performance. Thus, SSE-TSR is a powerful bioinformatics tool that improves protein classification and understanding of protein function and interaction.
Bias Neutralization Framework: Measuring Fairness in Large Language Models with Bias Intelligence Quotient (BiQ)
Apr 28, 2024Abstract:The burgeoning influence of Large Language Models (LLMs) in shaping public discourse and decision-making underscores the imperative to address inherent biases within these AI systems. In the wake of AI's expansive integration across sectors, addressing racial bias in LLMs has never been more critical. This paper introduces a novel framework called Comprehensive Bias Neutralization Framework (CBNF) which embodies an innovative approach to quantifying and mitigating biases within LLMs. Our framework combines the Large Language Model Bias Index (LLMBI) [Oketunji, A., Anas, M., Saina, D., (2023)] and Bias removaL with No Demographics (BLIND) [Orgad, H., Belinkov, Y. (2023)] methodologies to create a new metric called Bias Intelligence Quotient (BiQ)which detects, measures, and mitigates racial bias in LLMs without reliance on demographic annotations. By introducing a new metric called BiQ that enhances LLMBI with additional fairness metrics, CBNF offers a multi-dimensional metric for bias assessment, underscoring the necessity of a nuanced approach to fairness in AI [Mehrabi et al., 2021]. This paper presents a detailed analysis of Latimer AI (a language model incrementally trained on black history and culture) in comparison to ChatGPT 3.5, illustrating Latimer AI's efficacy in detecting racial, cultural, and gender biases through targeted training and refined bias mitigation strategies [Latimer & Bender, 2023].
Device-aware inference operations in SONOS nonvolatile memory arrays
Apr 02, 2020
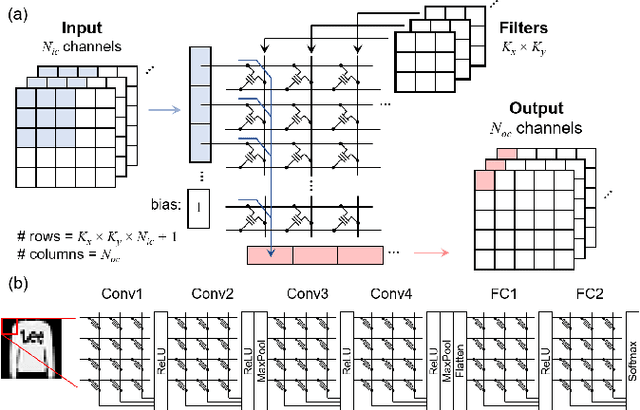


Abstract:Non-volatile memory arrays can deploy pre-trained neural network models for edge inference. However, these systems are affected by device-level noise and retention issues. Here, we examine damage caused by these effects, introduce a mitigation strategy, and demonstrate its use in fabricated array of SONOS (Silicon-Oxide-Nitride-Oxide-Silicon) devices. On MNIST, fashion-MNIST, and CIFAR-10 tasks, our approach increases resilience to synaptic noise and drift. We also show strong performance can be realized with ADCs of 5-8 bits precision.
Deep Multi-view Learning to Rank
Jan 31, 2018



Abstract:We study the problem of learning to rank from multiple sources. Though multi-view learning and learning to rank have been studied extensively leading to a wide range of applications, multi-view learning to rank as a synergy of both topics has received little attention. The aim of the paper is to propose a composite ranking method while keeping a close correlation with the individual rankings simultaneously. We propose a multi-objective solution to ranking by capturing the information of the feature mapping from both within each view as well as across views using autoencoder-like networks. Moreover, a novel end-to-end solution is introduced to enhance the joint ranking with minimum view-specific ranking loss, so that we can achieve the maximum global view agreements within a single optimization process. The proposed method is validated on a wide variety of ranking problems, including university ranking, multi-view lingual text ranking and image data ranking, providing superior results.
Multiple Kernel Learning and Automatic Subspace Relevance Determination for High-dimensional Neuroimaging Data
Jun 02, 2017



Abstract:Alzheimer's disease is a major cause of dementia. Its diagnosis requires accurate biomarkers that are sensitive to disease stages. In this respect, we regard probabilistic classification as a method of designing a probabilistic biomarker for disease staging. Probabilistic biomarkers naturally support the interpretation of decisions and evaluation of uncertainty associated with them. In this paper, we obtain probabilistic biomarkers via Gaussian Processes. Gaussian Processes enable probabilistic kernel machines that offer flexible means to accomplish Multiple Kernel Learning. Exploiting this flexibility, we propose a new variation of Automatic Relevance Determination and tackle the challenges of high dimensionality through multiple kernels. Our research results demonstrate that the Gaussian Process models are competitive with or better than the well-known Support Vector Machine in terms of classification performance even in the cases of single kernel learning. Extending the basic scheme towards the Multiple Kernel Learning, we improve the efficacy of the Gaussian Process models and their interpretability in terms of the known anatomical correlates of the disease. For instance, the disease pathology starts in and around the hippocampus and entorhinal cortex. Through the use of Gaussian Processes and Multiple Kernel Learning, we have automatically and efficiently determined those portions of neuroimaging data. In addition to their interpretability, our Gaussian Process models are competitive with recent deep learning solutions under similar settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge