Tri Minh Nguyen
Efficient Symmetry-Aware Materials Generation via Hierarchical Generative Flow Networks
Nov 06, 2024



Abstract:Discovering new solid-state materials requires rapidly exploring the vast space of crystal structures and locating stable regions. Generating stable materials with desired properties and compositions is extremely difficult as we search for very small isolated pockets in the exponentially many possibilities, considering elements from the periodic table and their 3D arrangements in crystal lattices. Materials discovery necessitates both optimized solution structures and diversity in the generated material structures. Existing methods struggle to explore large material spaces and generate diverse samples with desired properties and requirements. We propose the Symmetry-aware Hierarchical Architecture for Flow-based Traversal (SHAFT), a novel generative model employing a hierarchical exploration strategy to efficiently exploit the symmetry of the materials space to generate crystal structures given desired properties. In particular, our model decomposes the exponentially large materials space into a hierarchy of subspaces consisting of symmetric space groups, lattice parameters, and atoms. We demonstrate that SHAFT significantly outperforms state-of-the-art iterative generative methods, such as Generative Flow Networks (GFlowNets) and Crystal Diffusion Variational AutoEncoders (CDVAE), in crystal structure generation tasks, achieving higher validity, diversity, and stability of generated structures optimized for target properties and requirements.
Learning to Discover Medicines
Feb 14, 2022Abstract:Discovering new medicines is the hallmark of human endeavor to live a better and longer life. Yet the pace of discovery has slowed down as we need to venture into more wildly unexplored biomedical space to find one that matches today's high standard. Modern AI-enabled by powerful computing, large biomedical databases, and breakthroughs in deep learning-offers a new hope to break this loop as AI is rapidly maturing, ready to make a huge impact in the area. In this paper we review recent advances in AI methodologies that aim to crack this challenge. We organize the vast and rapidly growing literature of AI for drug discovery into three relatively stable sub-areas: (a) representation learning over molecular sequences and geometric graphs; (b) data-driven reasoning where we predict molecular properties and their binding, optimize existing compounds, generate de novo molecules, and plan the synthesis of target molecules; and (c) knowledge-based reasoning where we discuss the construction and reasoning over biomedical knowledge graphs. We will also identify open challenges and chart possible research directions for the years to come.
Mitigating cold start problems in drug-target affinity prediction with interaction knowledge transferring
Jan 16, 2022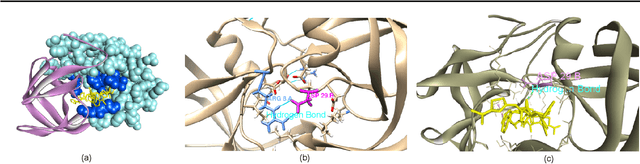
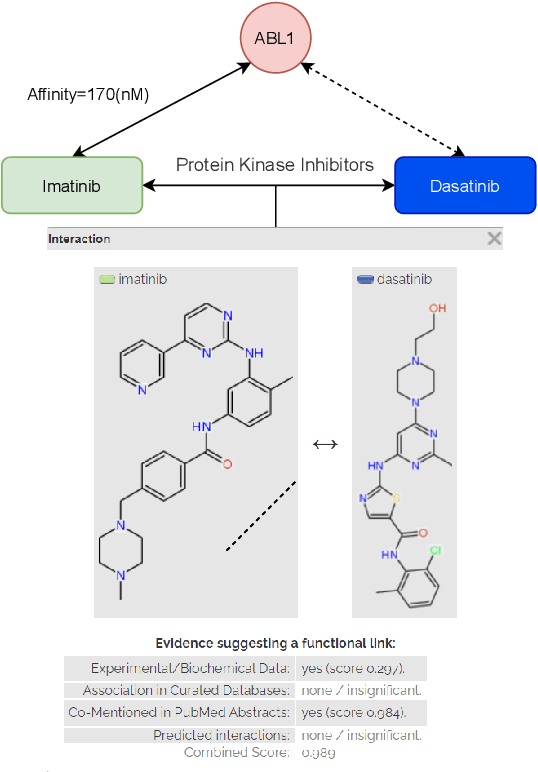
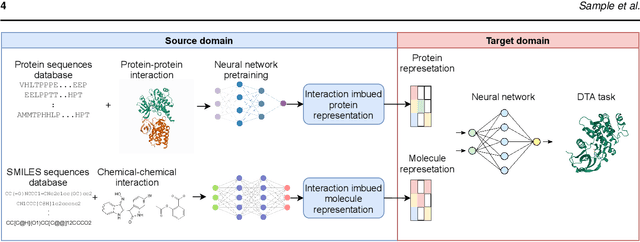
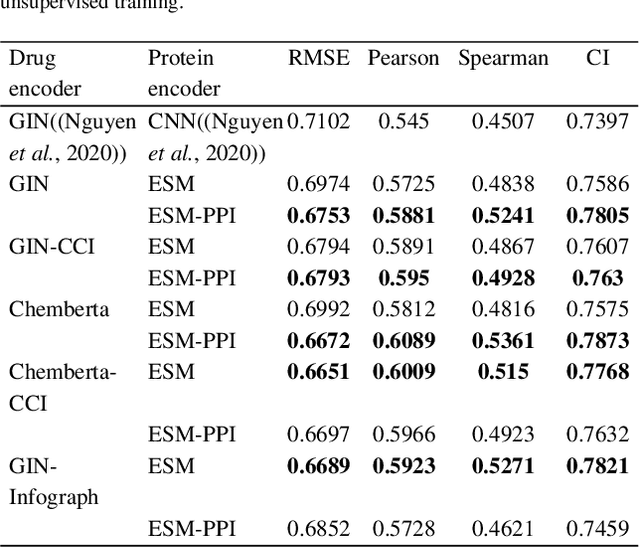
Abstract:Motivation: Predicting the drug-target interaction is crucial for drug discovery as well as drug repurposing. Machine learning is commonly used in drug-target affinity (DTA) problem. However, machine learning model faces the cold-start problem where the model performance drops when predicting the interaction of a novel drug or target. Previous works try to solve the cold start problem by learning the drug or target representation using unsupervised learning. While the drug or target representation can be learned in an unsupervised manner, it still lacks the interaction information, which is critical in drug-target interaction. Results: To incorporate the interaction information into the drug and protein interaction, we proposed using transfer learning from chemical-chemical interaction (CCI) and protein-protein interaction (PPI) task to drug-target interaction task. The representation learned by CCI and PPI tasks can be transferred smoothly to the DTA task due to the similar nature of the tasks. The result on the drug-target affinity datasets shows that our proposed method has advantages compared to other pretraining methods in the DTA task.
Counterfactual Explanation with Multi-Agent Reinforcement Learning for Drug Target Prediction
Mar 24, 2021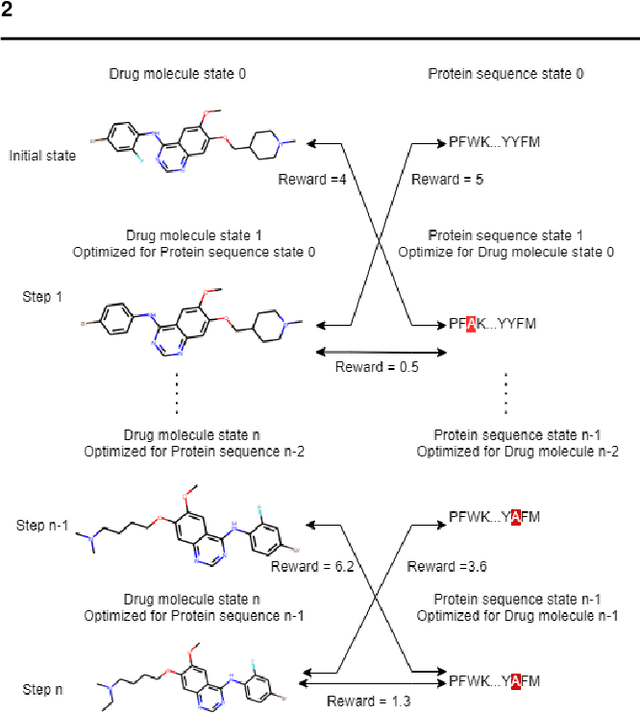
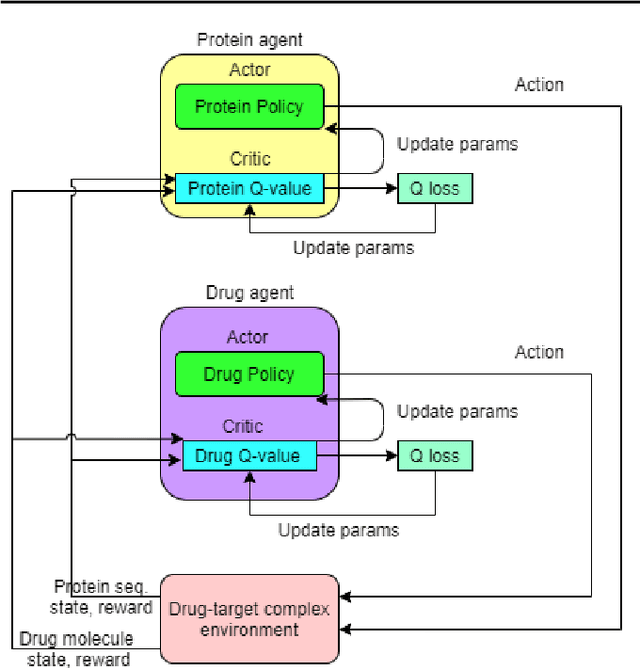
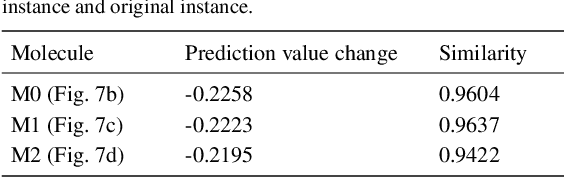
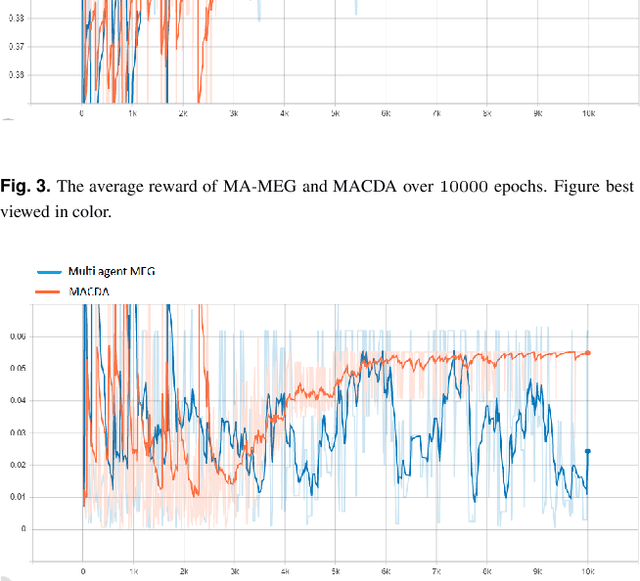
Abstract:Motivation: Several accurate deep learning models have been proposed to predict drug-target affinity (DTA). However, all of these models are black box hence are difficult to interpret and verify its result, and thus risking acceptance. Explanation is necessary to allow the DTA model more trustworthy. Explanation with counterfactual provides human-understandable examples. Most counterfactual explanation methods only operate on single input data, which are in tabular or continuous forms. In contrast, the DTA model has two discrete inputs. It is challenging for the counterfactual generation framework to optimize both discrete inputs at the same time. Results: We propose a multi-agent reinforcement learning framework, Multi-Agent Counterfactual Drug-target binding Affinity (MACDA), to generate counterfactual explanations for the drug-protein complex. Our proposed framework provides human-interpretable counterfactual instances while optimizing both the input drug and target for counterfactual generation at the same time. The result on the Davis dataset shows the advantages of the proposed MACDA framework compared with previous works.
GEFA: Early Fusion Approach in Drug-Target Affinity Prediction
Sep 28, 2020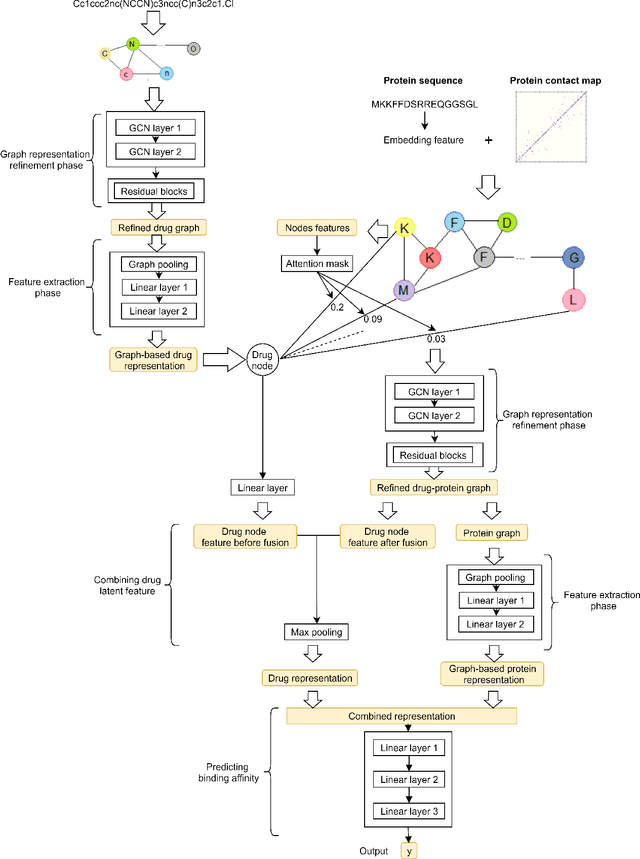
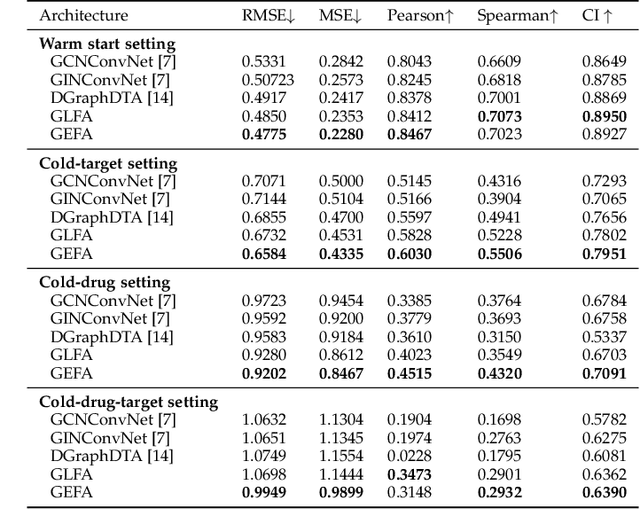
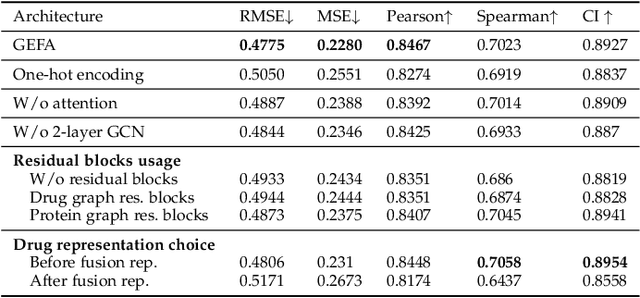

Abstract:Predicting the interaction between a compound and a target is crucial for rapid drug repurposing. Deep learning has been successfully applied in drug-target affinity (DTA) problem. However, previous deep learning-based methods ignore modeling the direct interactions between drug and protein residues. This would lead to inaccurate learning of target representation which may change due to the drug binding effects. In addition, previous DTA methods learn protein representation solely based on a small number of protein sequences in DTA datasets while neglecting the use of proteins outside of the DTA datasets. We propose GEFA (Graph Early Fusion Affinity), a novel graph-in-graph neural network with attention mechanism to address the changes in target representation because of the binding effects. Specifically, a drug is modeled as a graph of atoms, which then serves as a node in a larger graph of residues-drug complex. The resulting model is an expressive deep nested graph neural network. We also use pre-trained protein representation powered by the recent effort of learning contextualized protein representation. The experiments are conducted under different settings to evaluate scenarios such as novel drugs or targets. The results demonstrate the effectiveness of the pre-trained protein embedding and the advantages our GEFA in modeling the nested graph for drug-target interaction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge