Thomas P Quinn
A Field Guide to Scientific XAI: Transparent and Interpretable Deep Learning for Bioinformatics Research
Oct 13, 2021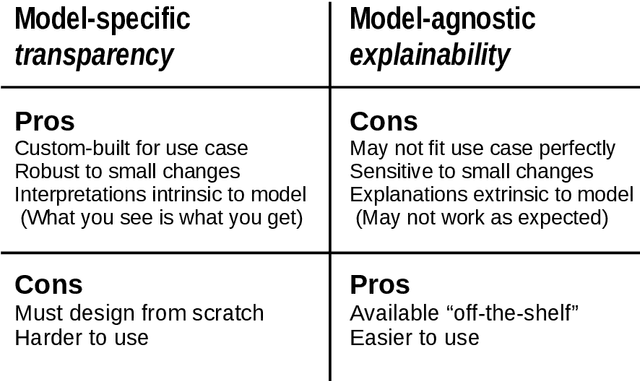
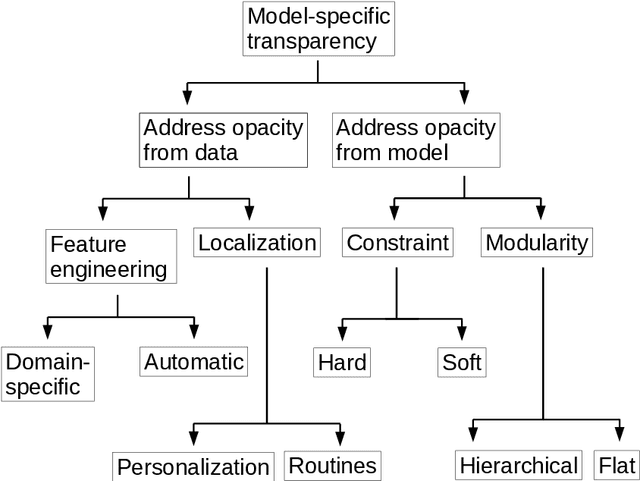
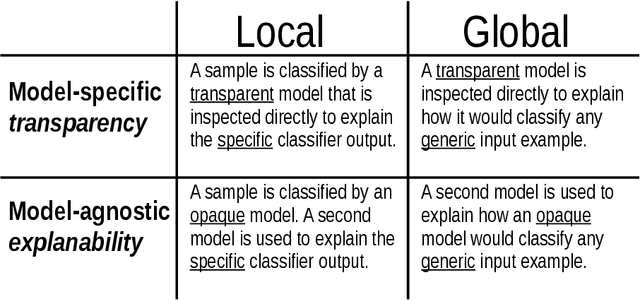
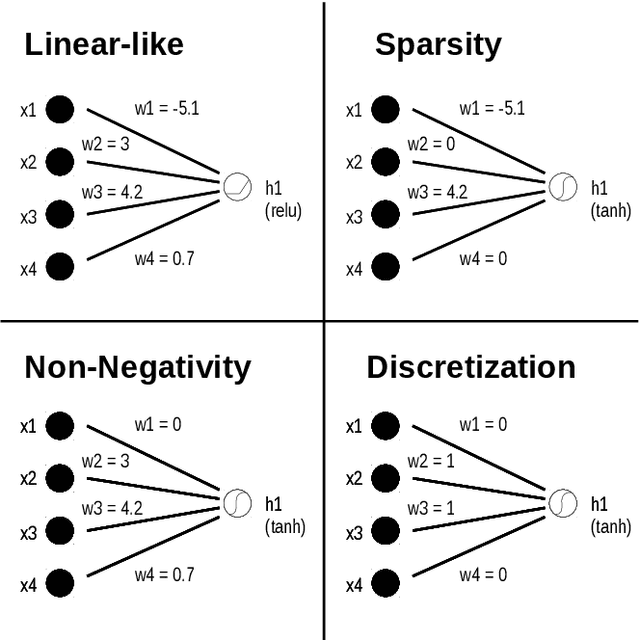
Abstract:Deep learning has become popular because of its potential to achieve high accuracy in prediction tasks. However, accuracy is not always the only goal of statistical modelling, especially for models developed as part of scientific research. Rather, many scientific models are developed to facilitate scientific discovery, by which we mean to abstract a human-understandable representation of the natural world. Unfortunately, the opacity of deep neural networks limit their role in scientific discovery, creating a new demand for models that are transparently interpretable. This article is a field guide to transparent model design. It provides a taxonomy of transparent model design concepts, a practical workflow for putting design concepts into practice, and a general template for reporting design choices. We hope this field guide will help researchers more effectively design transparently interpretable models, and thus enable them to use deep learning for scientific discovery.
Readying Medical Students for Medical AI: The Need to Embed AI Ethics Education
Sep 07, 2021
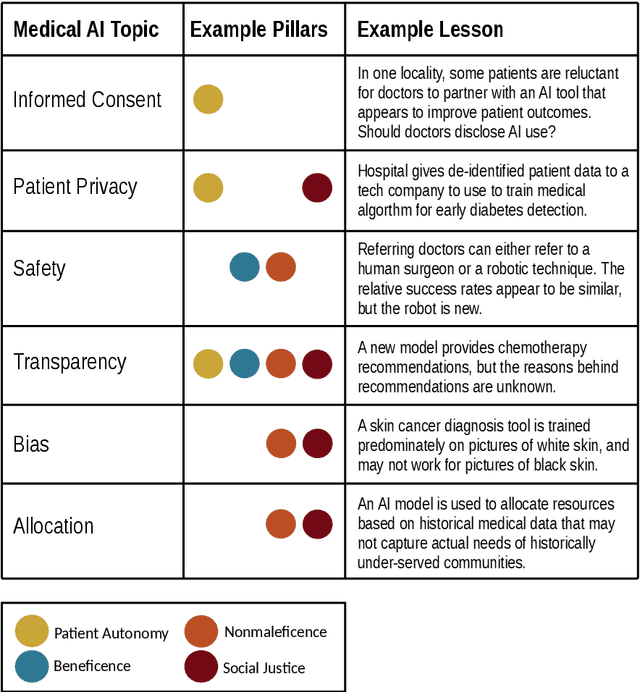
Abstract:Medical students will almost inevitably encounter powerful medical AI systems early in their careers. Yet, contemporary medical education does not adequately equip students with the basic clinical proficiency in medical AI needed to use these tools safely and effectively. Education reform is urgently needed, but not easily implemented, largely due to an already jam-packed medical curricula. In this article, we propose an education reform framework as an effective and efficient solution, which we call the Embedded AI Ethics Education Framework. Unlike other calls for education reform to accommodate AI teaching that are more radical in scope, our framework is modest and incremental. It leverages existing bioethics or medical ethics curricula to develop and deliver content on the ethical issues associated with medical AI, especially the harms of technology misuse, disuse, and abuse that affect the risk-benefit analyses at the heart of healthcare. In doing so, the framework provides a simple tool for going beyond the "What?" and the "Why?" of medical AI ethics education, to answer the "How?", giving universities, course directors, and/or professors a broad road-map for equipping their students with the necessary clinical proficiency in medical AI.
Counterfactual Explanation with Multi-Agent Reinforcement Learning for Drug Target Prediction
Mar 24, 2021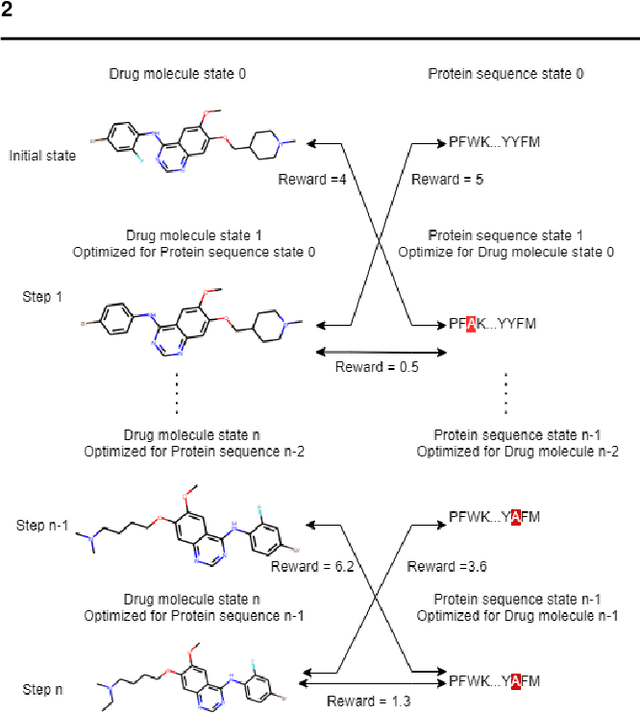
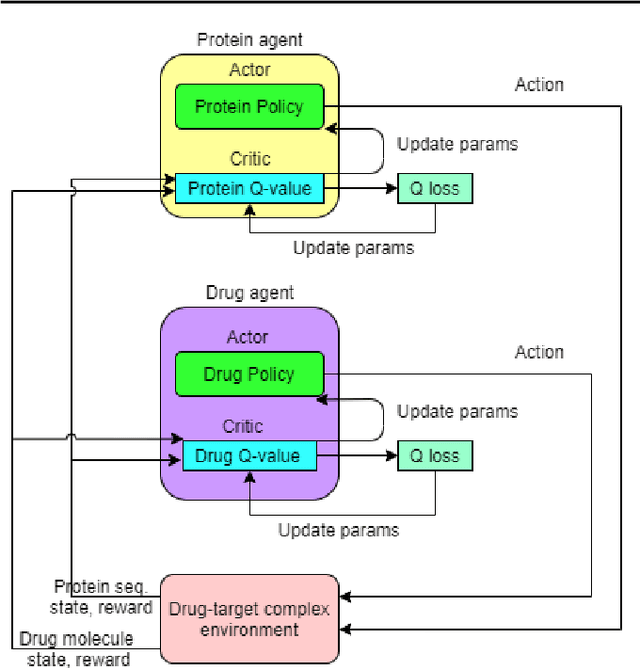
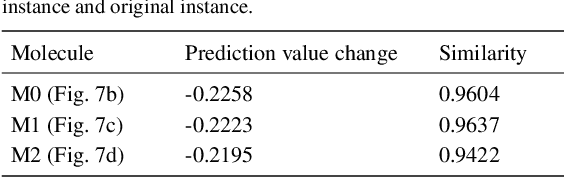
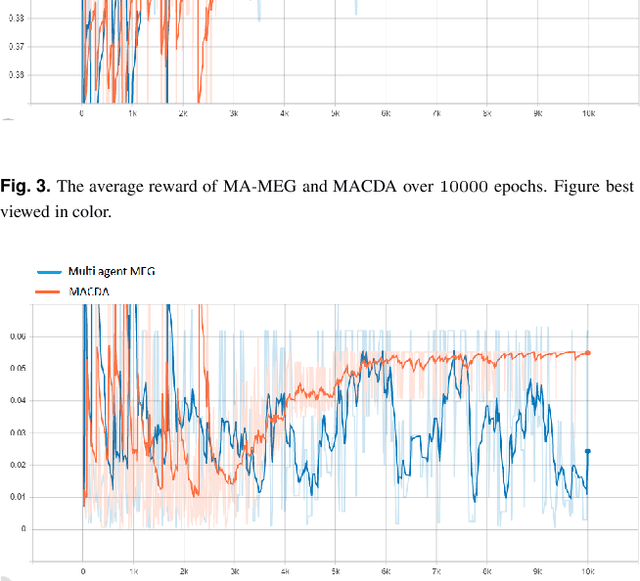
Abstract:Motivation: Several accurate deep learning models have been proposed to predict drug-target affinity (DTA). However, all of these models are black box hence are difficult to interpret and verify its result, and thus risking acceptance. Explanation is necessary to allow the DTA model more trustworthy. Explanation with counterfactual provides human-understandable examples. Most counterfactual explanation methods only operate on single input data, which are in tabular or continuous forms. In contrast, the DTA model has two discrete inputs. It is challenging for the counterfactual generation framework to optimize both discrete inputs at the same time. Results: We propose a multi-agent reinforcement learning framework, Multi-Agent Counterfactual Drug-target binding Affinity (MACDA), to generate counterfactual explanations for the drug-protein complex. Our proposed framework provides human-interpretable counterfactual instances while optimizing both the input drug and target for counterfactual generation at the same time. The result on the Davis dataset shows the advantages of the proposed MACDA framework compared with previous works.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge