Todd Hollon
Towards Scalable Language-Image Pre-training for 3D Medical Imaging
May 28, 2025Abstract:Language-image pre-training has demonstrated strong performance in 2D medical imaging, but its success in 3D modalities such as CT and MRI remains limited due to the high computational demands of volumetric data, which pose a significant barrier to training on large-scale, uncurated clinical studies. In this study, we introduce Hierarchical attention for Language-Image Pre-training (HLIP), a scalable pre-training framework for 3D medical imaging. HLIP adopts a lightweight hierarchical attention mechanism inspired by the natural hierarchy of radiology data: slice, scan, and study. This mechanism exhibits strong generalizability, e.g., +4.3% macro AUC on the Rad-ChestCT benchmark when pre-trained on CT-RATE. Moreover, the computational efficiency of HLIP enables direct training on uncurated datasets. Trained on 220K patients with 3.13 million scans for brain MRI and 240K patients with 1.44 million scans for head CT, HLIP achieves state-of-the-art performance, e.g., +32.4% balanced ACC on the proposed publicly available brain MRI benchmark Pub-Brain-5; +1.4% and +6.9% macro AUC on head CT benchmarks RSNA and CQ500, respectively. These results demonstrate that, with HLIP, directly pre-training on uncurated clinical datasets is a scalable and effective direction for language-image pre-training in 3D medical imaging. The code is available at https://github.com/Zch0414/hlip
Repurposing the scientific literature with vision-language models
Feb 26, 2025Abstract:Research in AI for Science often focuses on using AI technologies to augment components of the scientific process, or in some cases, the entire scientific method; how about AI for scientific publications? Peer-reviewed journals are foundational repositories of specialized knowledge, written in discipline-specific language that differs from general Internet content used to train most large language models (LLMs) and vision-language models (VLMs). We hypothesized that by combining a family of scientific journals with generative AI models, we could invent novel tools for scientific communication, education, and clinical care. We converted 23,000 articles from Neurosurgery Publications into a multimodal database - NeuroPubs - of 134 million words and 78,000 image-caption pairs to develop six datasets for building AI models. We showed that the content of NeuroPubs uniquely represents neurosurgery-specific clinical contexts compared with broader datasets and PubMed. For publishing, we employed generalist VLMs to automatically generate graphical abstracts from articles. Editorial board members rated 70% of these as ready for publication without further edits. For education, we generated 89,587 test questions in the style of the ABNS written board exam, which trainee and faculty neurosurgeons found indistinguishable from genuine examples 54% of the time. We used these questions alongside a curriculum learning process to track knowledge acquisition while training our 34 billion-parameter VLM (CNS-Obsidian). In a blinded, randomized controlled trial, we demonstrated the non-inferiority of CNS-Obsidian to GPT-4o (p = 0.1154) as a diagnostic copilot for a neurosurgical service. Our findings lay a novel foundation for AI with Science and establish a framework to elevate scientific communication using state-of-the-art generative artificial intelligence while maintaining rigorous quality standards.
Super-resolution of biomedical volumes with 2D supervision
Apr 15, 2024



Abstract:Volumetric biomedical microscopy has the potential to increase the diagnostic information extracted from clinical tissue specimens and improve the diagnostic accuracy of both human pathologists and computational pathology models. Unfortunately, barriers to integrating 3-dimensional (3D) volumetric microscopy into clinical medicine include long imaging times, poor depth / z-axis resolution, and an insufficient amount of high-quality volumetric data. Leveraging the abundance of high-resolution 2D microscopy data, we introduce masked slice diffusion for super-resolution (MSDSR), which exploits the inherent equivalence in the data-generating distribution across all spatial dimensions of biological specimens. This intrinsic characteristic allows for super-resolution models trained on high-resolution images from one plane (e.g., XY) to effectively generalize to others (XZ, YZ), overcoming the traditional dependency on orientation. We focus on the application of MSDSR to stimulated Raman histology (SRH), an optical imaging modality for biological specimen analysis and intraoperative diagnosis, characterized by its rapid acquisition of high-resolution 2D images but slow and costly optical z-sectioning. To evaluate MSDSR's efficacy, we introduce a new performance metric, SliceFID, and demonstrate MSDSR's superior performance over baseline models through extensive evaluations. Our findings reveal that MSDSR not only significantly enhances the quality and resolution of 3D volumetric data, but also addresses major obstacles hindering the broader application of 3D volumetric microscopy in clinical diagnostics and biomedical research.
Contrastive Representation Learning for Rapid Intraoperative Diagnosis of Skull Base Tumors Imaged Using Stimulated Raman Histology
Aug 08, 2021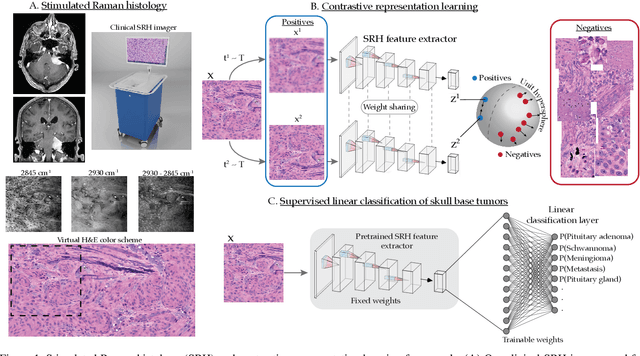
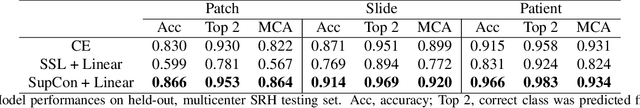
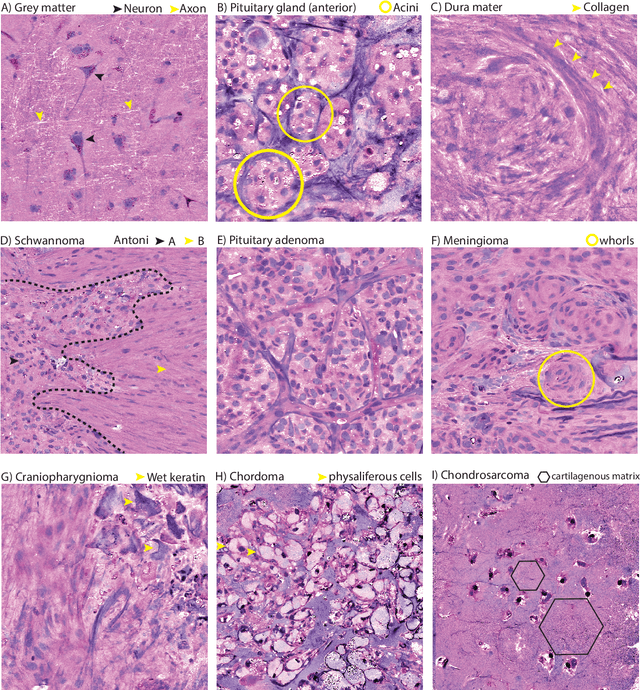
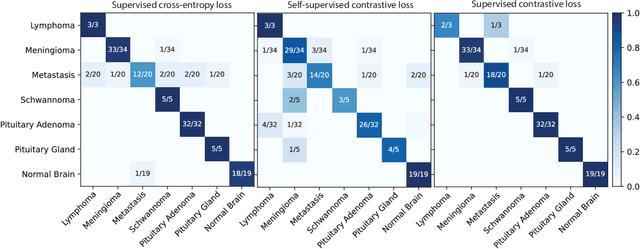
Abstract:Background: Accurate diagnosis of skull base tumors is essential for providing personalized surgical treatment strategies. Intraoperative diagnosis can be challenging due to tumor diversity and lack of intraoperative pathology resources. Objective: To develop an independent and parallel intraoperative pathology workflow that can provide rapid and accurate skull base tumor diagnoses using label-free optical imaging and artificial intelligence (AI). Method: We used a fiber laser-based, label-free, non-consumptive, high-resolution microscopy method ($<$ 60 sec per 1 $\times$ 1 mm$^\text{2}$), called stimulated Raman histology (SRH), to image a consecutive, multicenter cohort of skull base tumor patients. SRH images were then used to train a convolutional neural network (CNN) model using three representation learning strategies: cross-entropy, self-supervised contrastive learning, and supervised contrastive learning. Our trained CNN models were tested on a held-out, multicenter SRH dataset. Results: SRH was able to image the diagnostic features of both benign and malignant skull base tumors. Of the three representation learning strategies, supervised contrastive learning most effectively learned the distinctive and diagnostic SRH image features for each of the skull base tumor types. In our multicenter testing set, cross-entropy achieved an overall diagnostic accuracy of 91.5%, self-supervised contrastive learning 83.9%, and supervised contrastive learning 96.6%. Our trained model was able to identify tumor-normal margins and detect regions of microscopic tumor infiltration in whole-slide SRH images. Conclusion: SRH with AI models trained using contrastive representation learning can provide rapid and accurate intraoperative diagnosis of skull base tumors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge