Daniel Alber
BPQA Dataset: Evaluating How Well Language Models Leverage Blood Pressures to Answer Biomedical Questions
Mar 06, 2025Abstract:Clinical measurements such as blood pressures and respiration rates are critical in diagnosing and monitoring patient outcomes. It is an important component of biomedical data, which can be used to train transformer-based language models (LMs) for improving healthcare delivery. It is, however, unclear whether LMs can effectively interpret and use clinical measurements. We investigate two questions: First, can LMs effectively leverage clinical measurements to answer related medical questions? Second, how to enhance an LM's performance on medical question-answering (QA) tasks that involve measurements? We performed a case study on blood pressure readings (BPs), a vital sign routinely monitored by medical professionals. We evaluated the performance of four LMs: BERT, BioBERT, MedAlpaca, and GPT-3.5, on our newly developed dataset, BPQA (Blood Pressure Question Answering). BPQA contains $100$ medical QA pairs that were verified by medical students and designed to rely on BPs . We found that GPT-3.5 and MedAlpaca (larger and medium sized LMs) benefit more from the inclusion of BPs than BERT and BioBERT (small sized LMs). Further, augmenting measurements with labels improves the performance of BioBERT and Medalpaca (domain specific LMs), suggesting that retrieval may be useful for improving domain-specific LMs.
Contrastive Representation Learning for Rapid Intraoperative Diagnosis of Skull Base Tumors Imaged Using Stimulated Raman Histology
Aug 08, 2021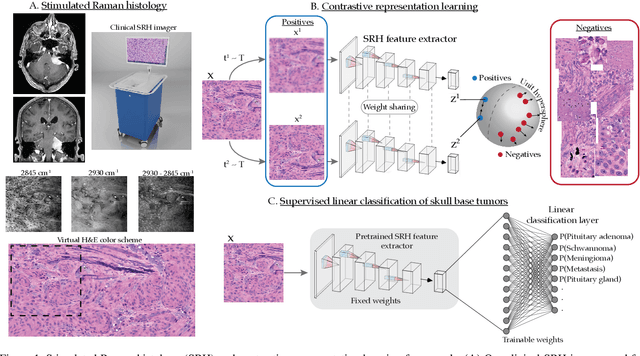
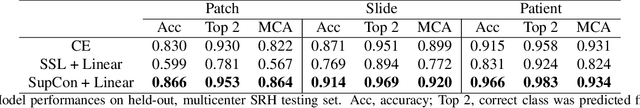
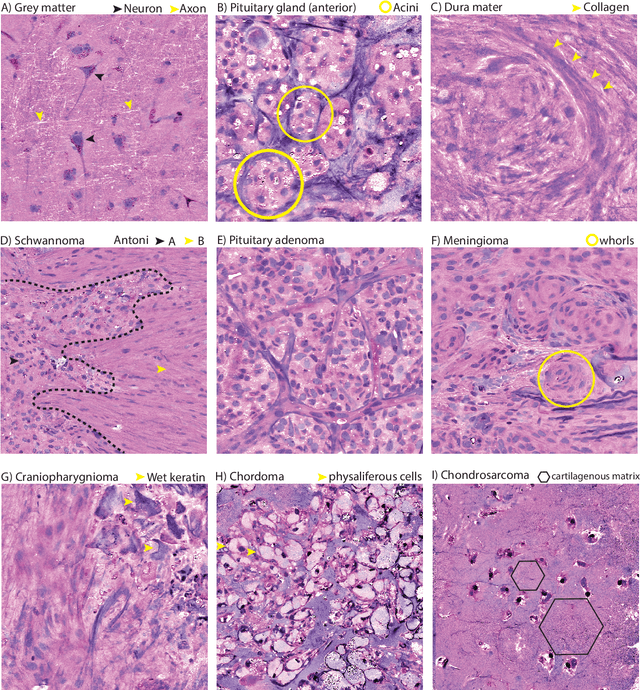
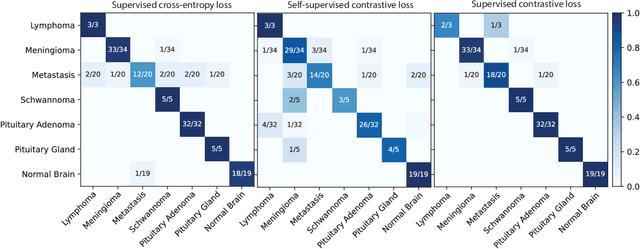
Abstract:Background: Accurate diagnosis of skull base tumors is essential for providing personalized surgical treatment strategies. Intraoperative diagnosis can be challenging due to tumor diversity and lack of intraoperative pathology resources. Objective: To develop an independent and parallel intraoperative pathology workflow that can provide rapid and accurate skull base tumor diagnoses using label-free optical imaging and artificial intelligence (AI). Method: We used a fiber laser-based, label-free, non-consumptive, high-resolution microscopy method ($<$ 60 sec per 1 $\times$ 1 mm$^\text{2}$), called stimulated Raman histology (SRH), to image a consecutive, multicenter cohort of skull base tumor patients. SRH images were then used to train a convolutional neural network (CNN) model using three representation learning strategies: cross-entropy, self-supervised contrastive learning, and supervised contrastive learning. Our trained CNN models were tested on a held-out, multicenter SRH dataset. Results: SRH was able to image the diagnostic features of both benign and malignant skull base tumors. Of the three representation learning strategies, supervised contrastive learning most effectively learned the distinctive and diagnostic SRH image features for each of the skull base tumor types. In our multicenter testing set, cross-entropy achieved an overall diagnostic accuracy of 91.5%, self-supervised contrastive learning 83.9%, and supervised contrastive learning 96.6%. Our trained model was able to identify tumor-normal margins and detect regions of microscopic tumor infiltration in whole-slide SRH images. Conclusion: SRH with AI models trained using contrastive representation learning can provide rapid and accurate intraoperative diagnosis of skull base tumors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge