Akhil Kondepudi
Learning complete and explainable visual representations from itemized text supervision
Dec 11, 2025Abstract:Training vision models with language supervision enables general and transferable representations. However, many visual domains, especially non-object-centric domains such as medical imaging and remote sensing, contain itemized text annotations: multiple text items describing distinct and semantically independent findings within a single image. Such supervision differs from standard multi-caption supervision, where captions are redundant or highly overlapping. Here, we introduce ItemizedCLIP, a framework for learning complete and explainable visual representations from itemized text supervision. ItemizedCLIP employs a cross-attention module to produce text item-conditioned visual embeddings and a set of tailored objectives that jointly enforce item independence (distinct regions for distinct items) and representation completeness (coverage of all items). Across four domains with naturally itemized text supervision (brain MRI, head CT, chest CT, remote sensing) and one additional synthetically itemized dataset, ItemizedCLIP achieves substantial improvements in zero-shot performance and fine-grained interpretability over baselines. The resulting ItemizedCLIP representations are semantically grounded, item-differentiable, complete, and visually interpretable. Our code is available at https://github.com/MLNeurosurg/ItemizedCLIP.
Towards Scalable Language-Image Pre-training for 3D Medical Imaging
May 28, 2025Abstract:Language-image pre-training has demonstrated strong performance in 2D medical imaging, but its success in 3D modalities such as CT and MRI remains limited due to the high computational demands of volumetric data, which pose a significant barrier to training on large-scale, uncurated clinical studies. In this study, we introduce Hierarchical attention for Language-Image Pre-training (HLIP), a scalable pre-training framework for 3D medical imaging. HLIP adopts a lightweight hierarchical attention mechanism inspired by the natural hierarchy of radiology data: slice, scan, and study. This mechanism exhibits strong generalizability, e.g., +4.3% macro AUC on the Rad-ChestCT benchmark when pre-trained on CT-RATE. Moreover, the computational efficiency of HLIP enables direct training on uncurated datasets. Trained on 220K patients with 3.13 million scans for brain MRI and 240K patients with 1.44 million scans for head CT, HLIP achieves state-of-the-art performance, e.g., +32.4% balanced ACC on the proposed publicly available brain MRI benchmark Pub-Brain-5; +1.4% and +6.9% macro AUC on head CT benchmarks RSNA and CQ500, respectively. These results demonstrate that, with HLIP, directly pre-training on uncurated clinical datasets is a scalable and effective direction for language-image pre-training in 3D medical imaging. The code is available at https://github.com/Zch0414/hlip
Super-resolution of biomedical volumes with 2D supervision
Apr 15, 2024



Abstract:Volumetric biomedical microscopy has the potential to increase the diagnostic information extracted from clinical tissue specimens and improve the diagnostic accuracy of both human pathologists and computational pathology models. Unfortunately, barriers to integrating 3-dimensional (3D) volumetric microscopy into clinical medicine include long imaging times, poor depth / z-axis resolution, and an insufficient amount of high-quality volumetric data. Leveraging the abundance of high-resolution 2D microscopy data, we introduce masked slice diffusion for super-resolution (MSDSR), which exploits the inherent equivalence in the data-generating distribution across all spatial dimensions of biological specimens. This intrinsic characteristic allows for super-resolution models trained on high-resolution images from one plane (e.g., XY) to effectively generalize to others (XZ, YZ), overcoming the traditional dependency on orientation. We focus on the application of MSDSR to stimulated Raman histology (SRH), an optical imaging modality for biological specimen analysis and intraoperative diagnosis, characterized by its rapid acquisition of high-resolution 2D images but slow and costly optical z-sectioning. To evaluate MSDSR's efficacy, we introduce a new performance metric, SliceFID, and demonstrate MSDSR's superior performance over baseline models through extensive evaluations. Our findings reveal that MSDSR not only significantly enhances the quality and resolution of 3D volumetric data, but also addresses major obstacles hindering the broader application of 3D volumetric microscopy in clinical diagnostics and biomedical research.
Step-Calibrated Diffusion for Biomedical Optical Image Restoration
Mar 26, 2024



Abstract:High-quality, high-resolution medical imaging is essential for clinical care. Raman-based biomedical optical imaging uses non-ionizing infrared radiation to evaluate human tissues in real time and is used for early cancer detection, brain tumor diagnosis, and intraoperative tissue analysis. Unfortunately, optical imaging is vulnerable to image degradation due to laser scattering and absorption, which can result in diagnostic errors and misguided treatment. Restoration of optical images is a challenging computer vision task because the sources of image degradation are multi-factorial, stochastic, and tissue-dependent, preventing a straightforward method to obtain paired low-quality/high-quality data. Here, we present Restorative Step-Calibrated Diffusion (RSCD), an unpaired image restoration method that views the image restoration problem as completing the finishing steps of a diffusion-based image generation task. RSCD uses a step calibrator model to dynamically determine the severity of image degradation and the number of steps required to complete the reverse diffusion process for image restoration. RSCD outperforms other widely used unpaired image restoration methods on both image quality and perceptual evaluation metrics for restoring optical images. Medical imaging experts consistently prefer images restored using RSCD in blinded comparison experiments and report minimal to no hallucinations. Finally, we show that RSCD improves performance on downstream clinical imaging tasks, including automated brain tumor diagnosis and deep tissue imaging. Our code is available at https://github.com/MLNeurosurg/restorative_step-calibrated_diffusion.
A self-supervised framework for learning whole slide representations
Feb 09, 2024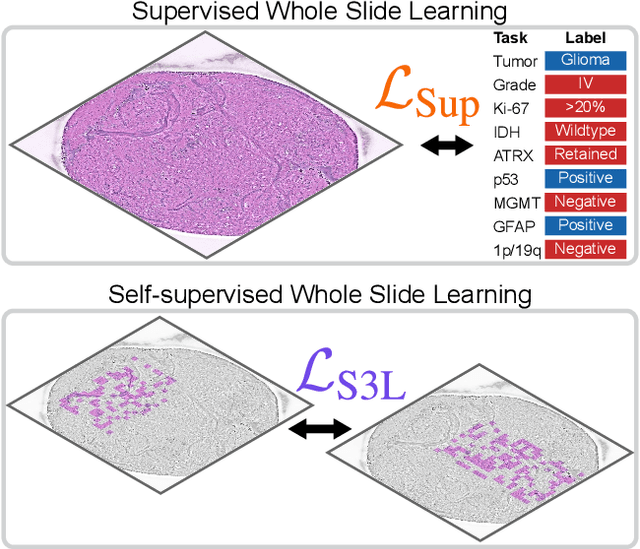
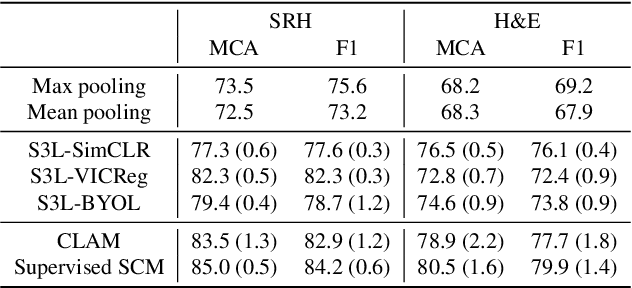
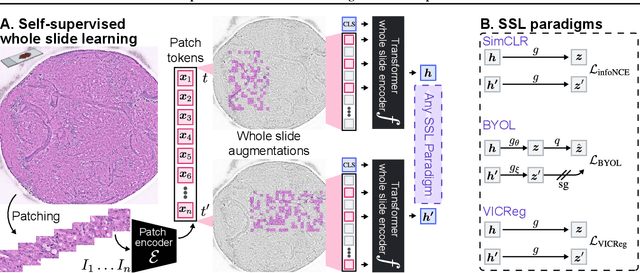
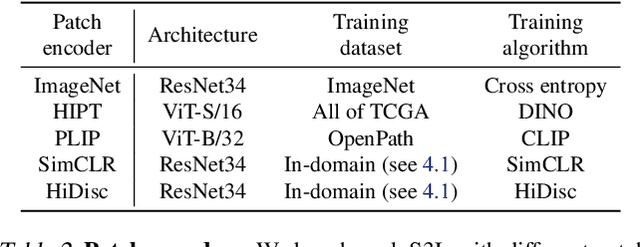
Abstract:Whole slide imaging is fundamental to biomedical microscopy and computational pathology. However, whole slide images (WSIs) present a complex computer vision challenge due to their gigapixel size, diverse histopathologic features, spatial heterogeneity, and limited/absent data annotations. These challenges highlight that supervised training alone can result in suboptimal whole slide representations. Self-supervised representation learning can achieve high-quality WSI visual feature learning for downstream diagnostic tasks, such as cancer diagnosis or molecular genetic prediction. Here, we present a general self-supervised whole slide learning (S3L) framework for gigapixel-scale self-supervision of WSIs. S3L combines data transformation strategies from transformer-based vision and language modeling into a single unified framework to generate paired views for self-supervision. S3L leverages the inherent regional heterogeneity, histologic feature variability, and information redundancy within WSIs to learn high-quality whole-slide representations. We benchmark S3L visual representations on two diagnostic tasks for two biomedical microscopy modalities. S3L significantly outperforms WSI baselines for cancer diagnosis and genetic mutation prediction. Additionally, S3L achieves good performance using both in-domain and out-of-distribution patch encoders, demonstrating good flexibility and generalizability.
Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging
Mar 23, 2023Abstract:Molecular classification has transformed the management of brain tumors by enabling more accurate prognostication and personalized treatment. However, timely molecular diagnostic testing for patients with brain tumors is limited, complicating surgical and adjuvant treatment and obstructing clinical trial enrollment. In this study, we developed DeepGlioma, a rapid ($< 90$ seconds), artificial-intelligence-based diagnostic screening system to streamline the molecular diagnosis of diffuse gliomas. DeepGlioma is trained using a multimodal dataset that includes stimulated Raman histology (SRH); a rapid, label-free, non-consumptive, optical imaging method; and large-scale, public genomic data. In a prospective, multicenter, international testing cohort of patients with diffuse glioma ($n=153$) who underwent real-time SRH imaging, we demonstrate that DeepGlioma can predict the molecular alterations used by the World Health Organization to define the adult-type diffuse glioma taxonomy (IDH mutation, 1p19q co-deletion and ATRX mutation), achieving a mean molecular classification accuracy of $93.3\pm 1.6\%$. Our results represent how artificial intelligence and optical histology can be used to provide a rapid and scalable adjunct to wet lab methods for the molecular screening of patients with diffuse glioma.
Hierarchical discriminative learning improves visual representations of biomedical microscopy
Mar 02, 2023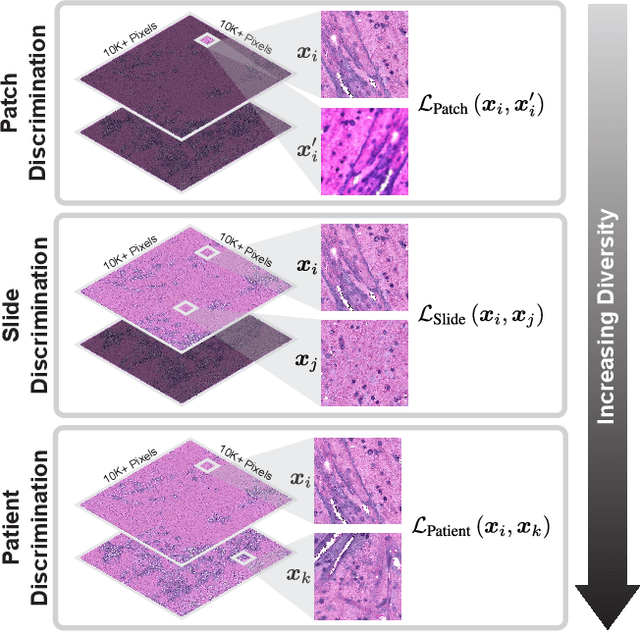
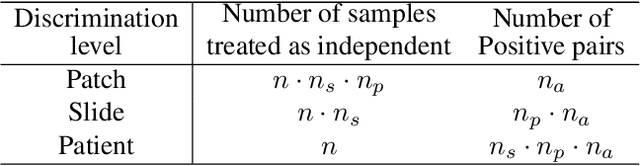
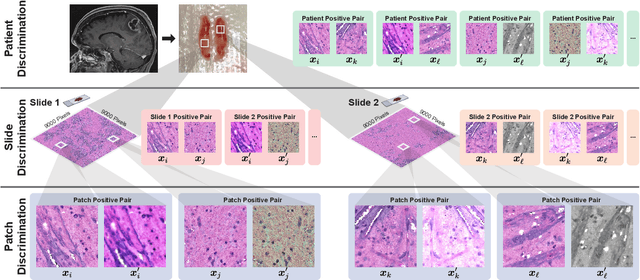
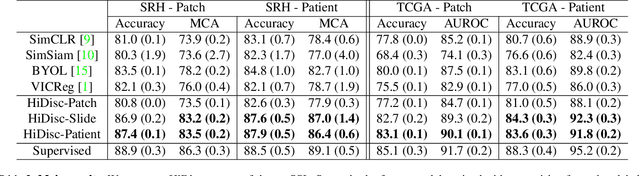
Abstract:Learning high-quality, self-supervised, visual representations is essential to advance the role of computer vision in biomedical microscopy and clinical medicine. Previous work has focused on self-supervised representation learning (SSL) methods developed for instance discrimination and applied them directly to image patches, or fields-of-view, sampled from gigapixel whole-slide images (WSIs) used for cancer diagnosis. However, this strategy is limited because it (1) assumes patches from the same patient are independent, (2) neglects the patient-slide-patch hierarchy of clinical biomedical microscopy, and (3) requires strong data augmentations that can degrade downstream performance. Importantly, sampled patches from WSIs of a patient's tumor are a diverse set of image examples that capture the same underlying cancer diagnosis. This motivated HiDisc, a data-driven method that leverages the inherent patient-slide-patch hierarchy of clinical biomedical microscopy to define a hierarchical discriminative learning task that implicitly learns features of the underlying diagnosis. HiDisc uses a self-supervised contrastive learning framework in which positive patch pairs are defined based on a common ancestry in the data hierarchy, and a unified patch, slide, and patient discriminative learning objective is used for visual SSL. We benchmark HiDisc visual representations on two vision tasks using two biomedical microscopy datasets, and demonstrate that (1) HiDisc pretraining outperforms current state-of-the-art self-supervised pretraining methods for cancer diagnosis and genetic mutation prediction, and (2) HiDisc learns high-quality visual representations using natural patch diversity without strong data augmentations.
OpenSRH: optimizing brain tumor surgery using intraoperative stimulated Raman histology
Jun 16, 2022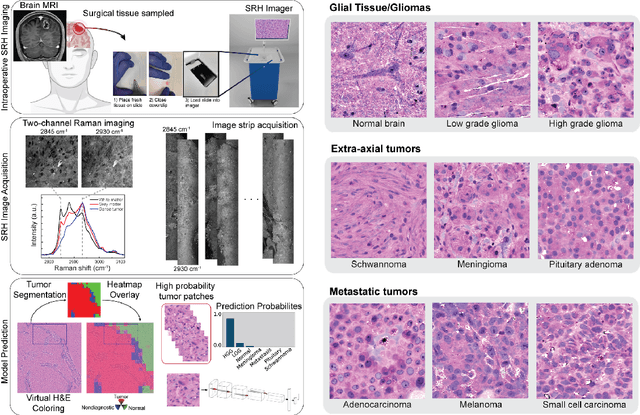
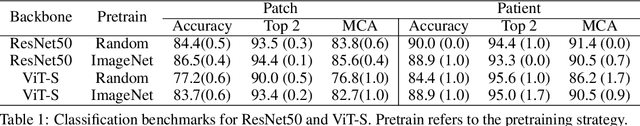
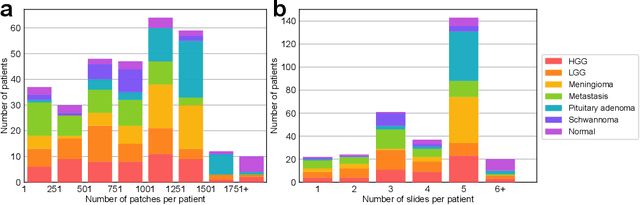
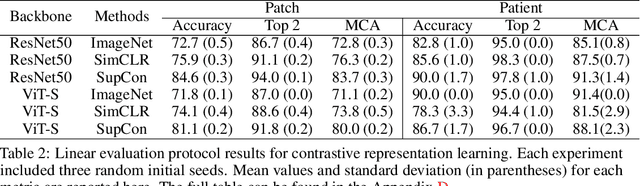
Abstract:Accurate intraoperative diagnosis is essential for providing safe and effective care during brain tumor surgery. Our standard-of-care diagnostic methods are time, resource, and labor intensive, which restricts access to optimal surgical treatments. To address these limitations, we propose an alternative workflow that combines stimulated Raman histology (SRH), a rapid optical imaging method, with deep learning-based automated interpretation of SRH images for intraoperative brain tumor diagnosis and real-time surgical decision support. Here, we present OpenSRH, the first public dataset of clinical SRH images from 300+ brain tumors patients and 1300+ unique whole slide optical images. OpenSRH contains data from the most common brain tumors diagnoses, full pathologic annotations, whole slide tumor segmentations, raw and processed optical imaging data for end-to-end model development and validation. We provide a framework for patch-based whole slide SRH classification and inference using weak (i.e. patient-level) diagnostic labels. Finally, we benchmark two computer vision tasks: multiclass histologic brain tumor classification and patch-based contrastive representation learning. We hope OpenSRH will facilitate the clinical translation of rapid optical imaging and real-time ML-based surgical decision support in order to improve the access, safety, and efficacy of cancer surgery in the era of precision medicine. Dataset access, code, and benchmarks are available at opensrh.mlins.org.
Contrastive Representation Learning for Rapid Intraoperative Diagnosis of Skull Base Tumors Imaged Using Stimulated Raman Histology
Aug 08, 2021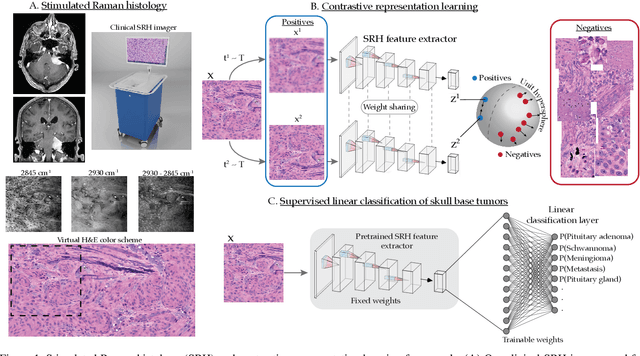
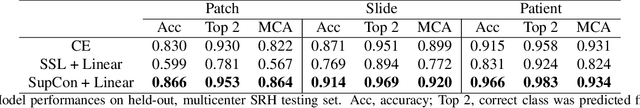
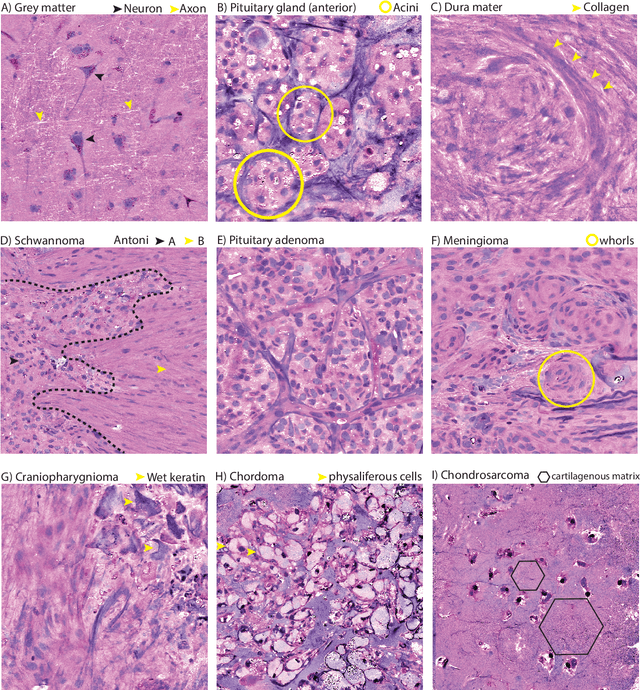
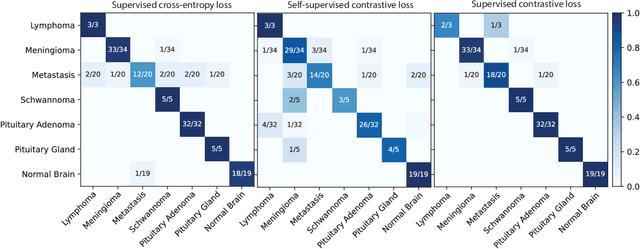
Abstract:Background: Accurate diagnosis of skull base tumors is essential for providing personalized surgical treatment strategies. Intraoperative diagnosis can be challenging due to tumor diversity and lack of intraoperative pathology resources. Objective: To develop an independent and parallel intraoperative pathology workflow that can provide rapid and accurate skull base tumor diagnoses using label-free optical imaging and artificial intelligence (AI). Method: We used a fiber laser-based, label-free, non-consumptive, high-resolution microscopy method ($<$ 60 sec per 1 $\times$ 1 mm$^\text{2}$), called stimulated Raman histology (SRH), to image a consecutive, multicenter cohort of skull base tumor patients. SRH images were then used to train a convolutional neural network (CNN) model using three representation learning strategies: cross-entropy, self-supervised contrastive learning, and supervised contrastive learning. Our trained CNN models were tested on a held-out, multicenter SRH dataset. Results: SRH was able to image the diagnostic features of both benign and malignant skull base tumors. Of the three representation learning strategies, supervised contrastive learning most effectively learned the distinctive and diagnostic SRH image features for each of the skull base tumor types. In our multicenter testing set, cross-entropy achieved an overall diagnostic accuracy of 91.5%, self-supervised contrastive learning 83.9%, and supervised contrastive learning 96.6%. Our trained model was able to identify tumor-normal margins and detect regions of microscopic tumor infiltration in whole-slide SRH images. Conclusion: SRH with AI models trained using contrastive representation learning can provide rapid and accurate intraoperative diagnosis of skull base tumors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge