Timothy Donovan
Coalitions of Large Language Models Increase the Robustness of AI Agents
Aug 02, 2024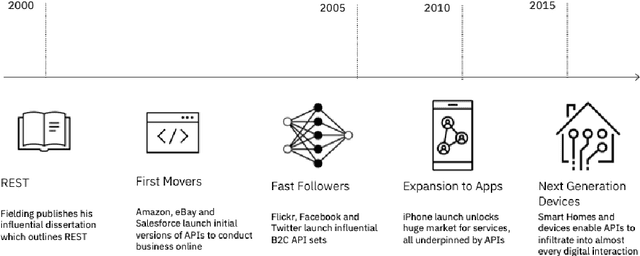
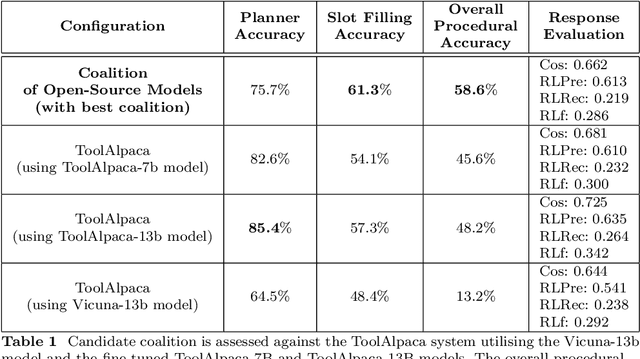
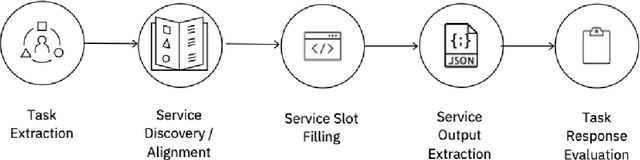
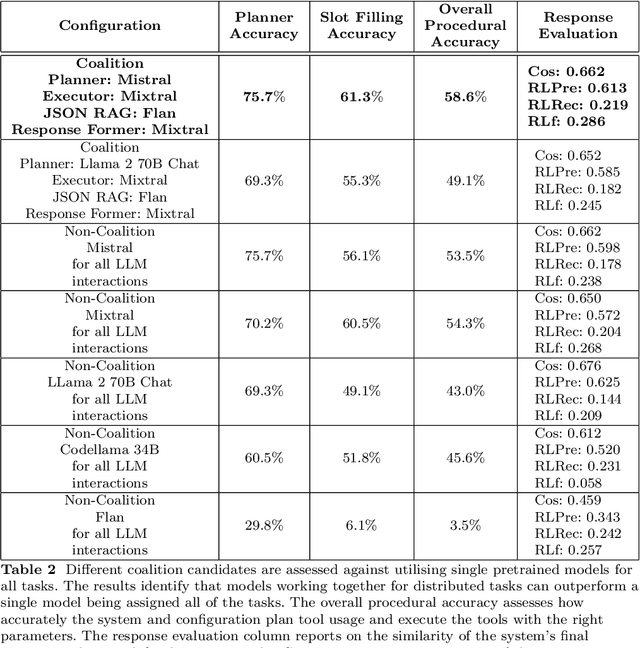
Abstract:The emergence of Large Language Models (LLMs) have fundamentally altered the way we interact with digital systems and have led to the pursuit of LLM powered AI agents to assist in daily workflows. LLMs, whilst powerful and capable of demonstrating some emergent properties, are not logical reasoners and often struggle to perform well at all sub-tasks carried out by an AI agent to plan and execute a workflow. While existing studies tackle this lack of proficiency by generalised pretraining at a huge scale or by specialised fine-tuning for tool use, we assess if a system comprising of a coalition of pretrained LLMs, each exhibiting specialised performance at individual sub-tasks, can match the performance of single model agents. The coalition of models approach showcases its potential for building robustness and reducing the operational costs of these AI agents by leveraging traits exhibited by specific models. Our findings demonstrate that fine-tuning can be mitigated by considering a coalition of pretrained models and believe that this approach can be applied to other non-agentic systems which utilise LLMs.
GT4SD: Generative Toolkit for Scientific Discovery
Jul 08, 2022
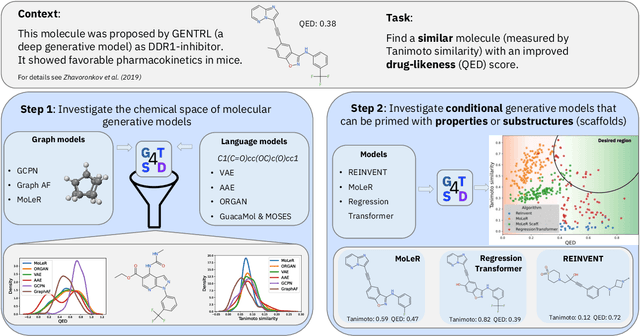
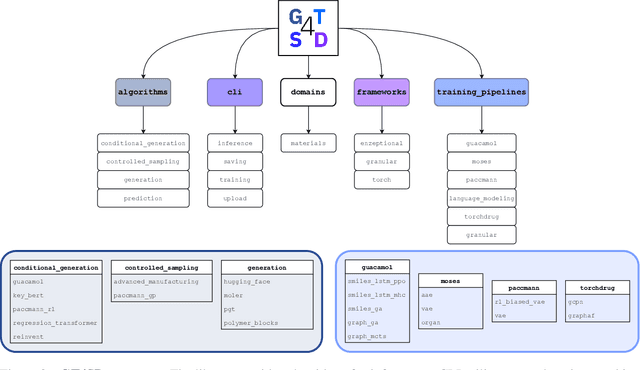
Abstract:With the growing availability of data within various scientific domains, generative models hold enormous potential to accelerate scientific discovery at every step of the scientific method. Perhaps their most valuable application lies in the speeding up of what has traditionally been the slowest and most challenging step of coming up with a hypothesis. Powerful representations are now being learned from large volumes of data to generate novel hypotheses, which is making a big impact on scientific discovery applications ranging from material design to drug discovery. The GT4SD (https://github.com/GT4SD/gt4sd-core) is an extensible open-source library that enables scientists, developers and researchers to train and use state-of-the-art generative models for hypothesis generation in scientific discovery. GT4SD supports a variety of uses of generative models across material science and drug discovery, including molecule discovery and design based on properties related to target proteins, omic profiles, scaffold distances, binding energies and more.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge