Tianye Niu
MulGT: Multi-task Graph-Transformer with Task-aware Knowledge Injection and Domain Knowledge-driven Pooling for Whole Slide Image Analysis
Feb 21, 2023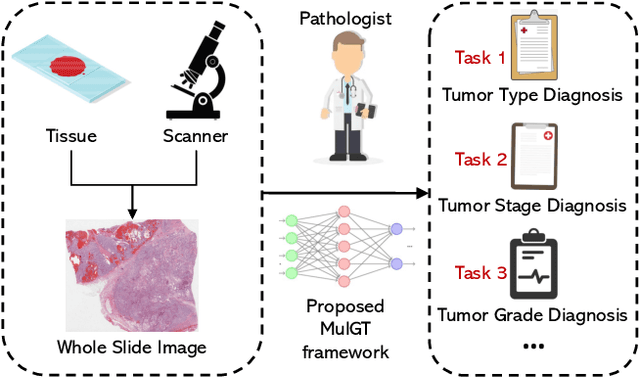
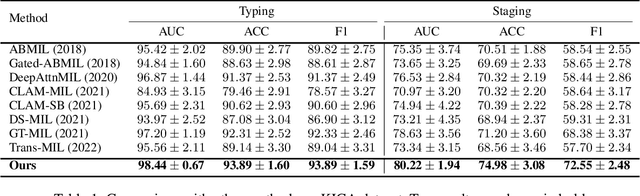
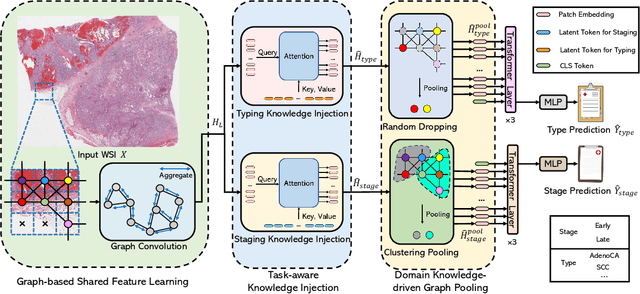
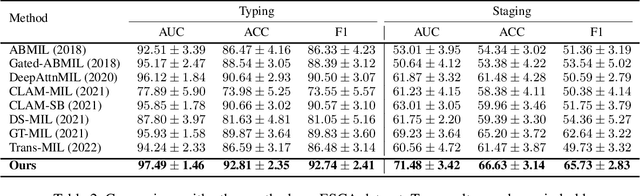
Abstract:Whole slide image (WSI) has been widely used to assist automated diagnosis under the deep learning fields. However, most previous works only discuss the SINGLE task setting which is not aligned with real clinical setting, where pathologists often conduct multiple diagnosis tasks simultaneously. Also, it is commonly recognized that the multi-task learning paradigm can improve learning efficiency by exploiting commonalities and differences across multiple tasks. To this end, we present a novel multi-task framework (i.e., MulGT) for WSI analysis by the specially designed Graph-Transformer equipped with Task-aware Knowledge Injection and Domain Knowledge-driven Graph Pooling modules. Basically, with the Graph Neural Network and Transformer as the building commons, our framework is able to learn task-agnostic low-level local information as well as task-specific high-level global representation. Considering that different tasks in WSI analysis depend on different features and properties, we also design a novel Task-aware Knowledge Injection module to transfer the task-shared graph embedding into task-specific feature spaces to learn more accurate representation for different tasks. Further, we elaborately design a novel Domain Knowledge-driven Graph Pooling module for each task to improve both the accuracy and robustness of different tasks by leveraging different diagnosis patterns of multiple tasks. We evaluated our method on two public WSI datasets from TCGA projects, i.e., esophageal carcinoma and kidney carcinoma. Experimental results show that our method outperforms single-task counterparts and the state-of-theart methods on both tumor typing and staging tasks.
ISA-Net: Improved spatial attention network for PET-CT tumor segmentation
Nov 04, 2022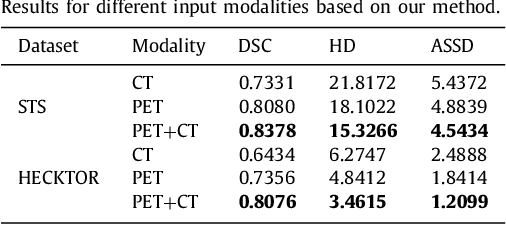
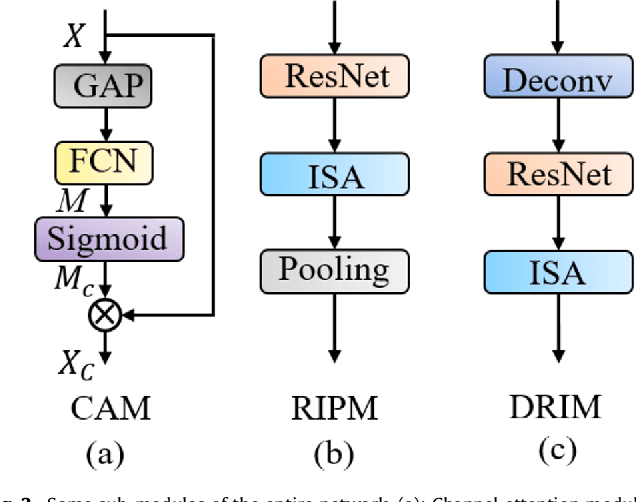
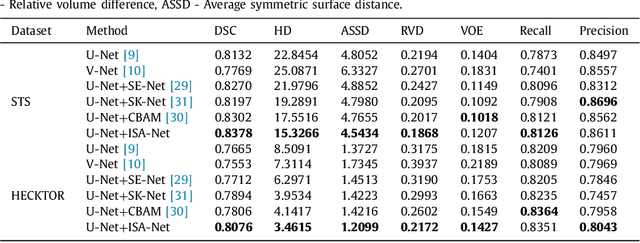
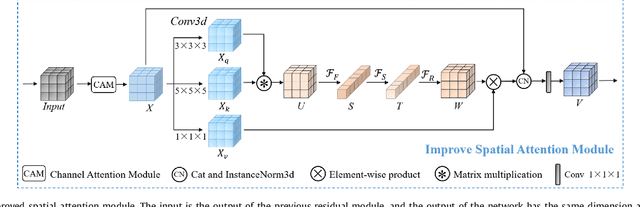
Abstract:Achieving accurate and automated tumor segmentation plays an important role in both clinical practice and radiomics research. Segmentation in medicine is now often performed manually by experts, which is a laborious, expensive and error-prone task. Manual annotation relies heavily on the experience and knowledge of these experts. In addition, there is much intra- and interobserver variation. Therefore, it is of great significance to develop a method that can automatically segment tumor target regions. In this paper, we propose a deep learning segmentation method based on multimodal positron emission tomography-computed tomography (PET-CT), which combines the high sensitivity of PET and the precise anatomical information of CT. We design an improved spatial attention network(ISA-Net) to increase the accuracy of PET or CT in detecting tumors, which uses multi-scale convolution operation to extract feature information and can highlight the tumor region location information and suppress the non-tumor region location information. In addition, our network uses dual-channel inputs in the coding stage and fuses them in the decoding stage, which can take advantage of the differences and complementarities between PET and CT. We validated the proposed ISA-Net method on two clinical datasets, a soft tissue sarcoma(STS) and a head and neck tumor(HECKTOR) dataset, and compared with other attention methods for tumor segmentation. The DSC score of 0.8378 on STS dataset and 0.8076 on HECKTOR dataset show that ISA-Net method achieves better segmentation performance and has better generalization. Conclusions: The method proposed in this paper is based on multi-modal medical image tumor segmentation, which can effectively utilize the difference and complementarity of different modes. The method can also be applied to other multi-modal data or single-modal data by proper adjustment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge