Stefan Gugler
How simple can you go? An off-the-shelf transformer approach to molecular dynamics
Mar 05, 2025Abstract:Most current neural networks for molecular dynamics (MD) include physical inductive biases, resulting in specialized and complex architectures. This is in contrast to most other machine learning domains, where specialist approaches are increasingly replaced by general-purpose architectures trained on vast datasets. In line with this trend, several recent studies have questioned the necessity of architectural features commonly found in MD models, such as built-in rotational equivariance or energy conservation. In this work, we contribute to the ongoing discussion by evaluating the performance of an MD model with as few specialized architectural features as possible. We present a recipe for MD using an Edge Transformer, an "off-the-shelf'' transformer architecture that has been minimally modified for the MD domain, termed MD-ET. Our model implements neither built-in equivariance nor energy conservation. We use a simple supervised pre-training scheme on $\sim$30 million molecular structures from the QCML database. Using this "off-the-shelf'' approach, we show state-of-the-art results on several benchmarks after fine-tuning for a small number of steps. Additionally, we examine the effects of being only approximately equivariant and energy conserving for MD simulations, proposing a novel method for distinguishing the errors resulting from non-equivariance from other sources of inaccuracies like numerical rounding errors. While our model exhibits runaway energy increases on larger structures, we show approximately energy-conserving NVE simulations for a range of small structures.
Enhancing Diffusion Models Efficiency by Disentangling Total-Variance and Signal-to-Noise Ratio
Feb 12, 2025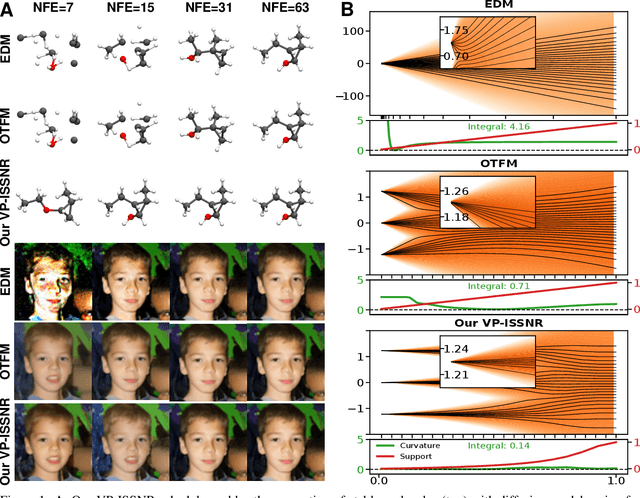
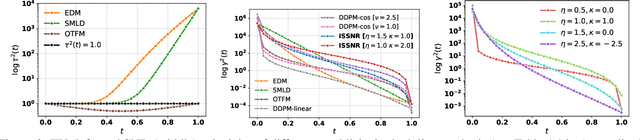
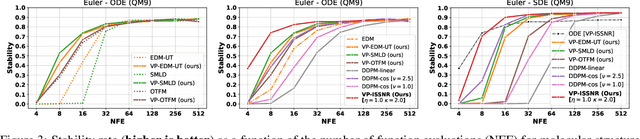
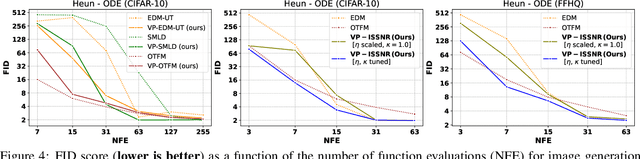
Abstract:The long sampling time of diffusion models remains a significant bottleneck, which can be mitigated by reducing the number of diffusion time steps. However, the quality of samples with fewer steps is highly dependent on the noise schedule, i.e., the specific manner in which noise is introduced and the signal is reduced at each step. Although prior work has improved upon the original variance-preserving and variance-exploding schedules, these approaches $\textit{passively}$ adjust the total variance, without direct control over it. In this work, we propose a novel total-variance/signal-to-noise-ratio disentangled (TV/SNR) framework, where TV and SNR can be controlled independently. Our approach reveals that different existing schedules, where the TV explodes exponentially, can be $\textit{improved}$ by setting a constant TV schedule while preserving the same SNR schedule. Furthermore, generalizing the SNR schedule of the optimal transport flow matching significantly improves the performance in molecular structure generation, achieving few step generation of stable molecules. A similar tendency is observed in image generation, where our approach with a uniform diffusion time grid performs comparably to the highly tailored EDM sampler.
Towards Symbolic XAI -- Explanation Through Human Understandable Logical Relationships Between Features
Aug 30, 2024Abstract:Explainable Artificial Intelligence (XAI) plays a crucial role in fostering transparency and trust in AI systems, where traditional XAI approaches typically offer one level of abstraction for explanations, often in the form of heatmaps highlighting single or multiple input features. However, we ask whether abstract reasoning or problem-solving strategies of a model may also be relevant, as these align more closely with how humans approach solutions to problems. We propose a framework, called Symbolic XAI, that attributes relevance to symbolic queries expressing logical relationships between input features, thereby capturing the abstract reasoning behind a model's predictions. The methodology is built upon a simple yet general multi-order decomposition of model predictions. This decomposition can be specified using higher-order propagation-based relevance methods, such as GNN-LRP, or perturbation-based explanation methods commonly used in XAI. The effectiveness of our framework is demonstrated in the domains of natural language processing (NLP), vision, and quantum chemistry (QC), where abstract symbolic domain knowledge is abundant and of significant interest to users. The Symbolic XAI framework provides an understanding of the model's decision-making process that is both flexible for customization by the user and human-readable through logical formulas.
Molecular relaxation by reverse diffusion with time step prediction
Apr 16, 2024



Abstract:Molecular relaxation, finding the equilibrium state of a non-equilibrium structure, is an essential component of computational chemistry to understand reactivity. Classical force field methods often rely on insufficient local energy minimization, while neural network force field models require large labeled datasets encompassing both equilibrium and non-equilibrium structures. As a remedy, we propose MoreRed, molecular relaxation by reverse diffusion, a conceptually novel and purely statistical approach where non-equilibrium structures are treated as noisy instances of their corresponding equilibrium states. To enable the denoising of arbitrarily noisy inputs via a generative diffusion model, we further introduce a novel diffusion time step predictor. Notably, MoreRed learns a simpler pseudo potential energy surface instead of the complex physical potential energy surface. It is trained on a significantly smaller, and thus computationally cheaper, dataset consisting of solely unlabeled equilibrium structures, avoiding the computation of non-equilibrium structures altogether. We compare MoreRed to classical force fields, equivariant neural network force fields trained on a large dataset of equilibrium and non-equilibrium data, as well as a semi-empirical tight-binding model. To assess this quantitatively, we evaluate the root-mean-square deviation between the found equilibrium structures and the reference equilibrium structures as well as their DFT energies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge