Khaled Kahouli
Control Variate Score Matching for Diffusion Models
Dec 23, 2025Abstract:Diffusion models offer a robust framework for sampling from unnormalized probability densities, which requires accurately estimating the score of the noise-perturbed target distribution. While the standard Denoising Score Identity (DSI) relies on data samples, access to the target energy function enables an alternative formulation via the Target Score Identity (TSI). However, these estimators face a fundamental variance trade-off: DSI exhibits high variance in low-noise regimes, whereas TSI suffers from high variance at high noise levels. In this work, we reconcile these approaches by unifying both estimators within the principled framework of control variates. We introduce the Control Variate Score Identity (CVSI), deriving an optimal, time-dependent control coefficient that theoretically guarantees variance minimization across the entire noise spectrum. We demonstrate that CVSI serves as a robust, low-variance plug-in estimator that significantly enhances sample efficiency in both data-free sampler learning and inference-time diffusion sampling.
Enhancing Diffusion Models Efficiency by Disentangling Total-Variance and Signal-to-Noise Ratio
Feb 12, 2025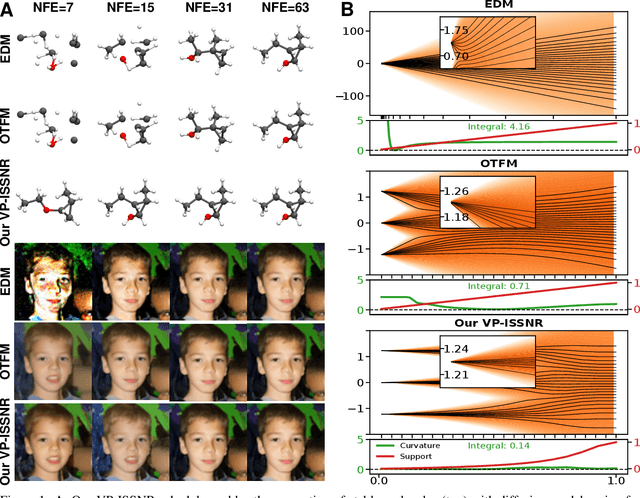
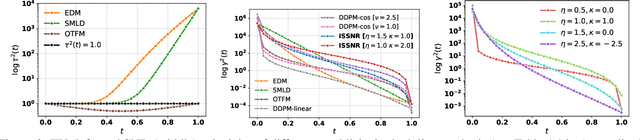
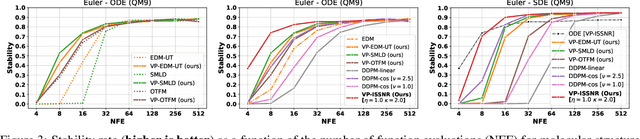
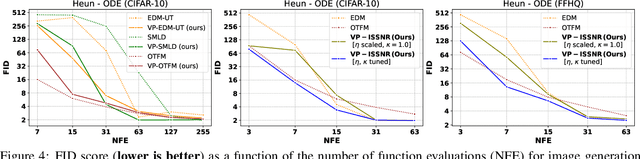
Abstract:The long sampling time of diffusion models remains a significant bottleneck, which can be mitigated by reducing the number of diffusion time steps. However, the quality of samples with fewer steps is highly dependent on the noise schedule, i.e., the specific manner in which noise is introduced and the signal is reduced at each step. Although prior work has improved upon the original variance-preserving and variance-exploding schedules, these approaches $\textit{passively}$ adjust the total variance, without direct control over it. In this work, we propose a novel total-variance/signal-to-noise-ratio disentangled (TV/SNR) framework, where TV and SNR can be controlled independently. Our approach reveals that different existing schedules, where the TV explodes exponentially, can be $\textit{improved}$ by setting a constant TV schedule while preserving the same SNR schedule. Furthermore, generalizing the SNR schedule of the optimal transport flow matching significantly improves the performance in molecular structure generation, achieving few step generation of stable molecules. A similar tendency is observed in image generation, where our approach with a uniform diffusion time grid performs comparably to the highly tailored EDM sampler.
Molecular relaxation by reverse diffusion with time step prediction
Apr 16, 2024



Abstract:Molecular relaxation, finding the equilibrium state of a non-equilibrium structure, is an essential component of computational chemistry to understand reactivity. Classical force field methods often rely on insufficient local energy minimization, while neural network force field models require large labeled datasets encompassing both equilibrium and non-equilibrium structures. As a remedy, we propose MoreRed, molecular relaxation by reverse diffusion, a conceptually novel and purely statistical approach where non-equilibrium structures are treated as noisy instances of their corresponding equilibrium states. To enable the denoising of arbitrarily noisy inputs via a generative diffusion model, we further introduce a novel diffusion time step predictor. Notably, MoreRed learns a simpler pseudo potential energy surface instead of the complex physical potential energy surface. It is trained on a significantly smaller, and thus computationally cheaper, dataset consisting of solely unlabeled equilibrium structures, avoiding the computation of non-equilibrium structures altogether. We compare MoreRed to classical force fields, equivariant neural network force fields trained on a large dataset of equilibrium and non-equilibrium data, as well as a semi-empirical tight-binding model. To assess this quantitatively, we evaluate the root-mean-square deviation between the found equilibrium structures and the reference equilibrium structures as well as their DFT energies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge