Stanisław Saganowski
Scaling Representation Learning from Ubiquitous ECG with State-Space Models
Sep 26, 2023Abstract:Ubiquitous sensing from wearable devices in the wild holds promise for enhancing human well-being, from diagnosing clinical conditions and measuring stress to building adaptive health promoting scaffolds. But the large volumes of data therein across heterogeneous contexts pose challenges for conventional supervised learning approaches. Representation Learning from biological signals is an emerging realm catalyzed by the recent advances in computational modeling and the abundance of publicly shared databases. The electrocardiogram (ECG) is the primary researched modality in this context, with applications in health monitoring, stress and affect estimation. Yet, most studies are limited by small-scale controlled data collection and over-parameterized architecture choices. We introduce \textbf{WildECG}, a pre-trained state-space model for representation learning from ECG signals. We train this model in a self-supervised manner with 275,000 10s ECG recordings collected in the wild and evaluate it on a range of downstream tasks. The proposed model is a robust backbone for ECG analysis, providing competitive performance on most of the tasks considered, while demonstrating efficacy in low-resource regimes. The code and pre-trained weights are shared publicly at https://github.com/klean2050/tiles_ecg_model.
Emotion Recognition Using Wearables: A Systematic Literature Review Work in progress
Jan 15, 2020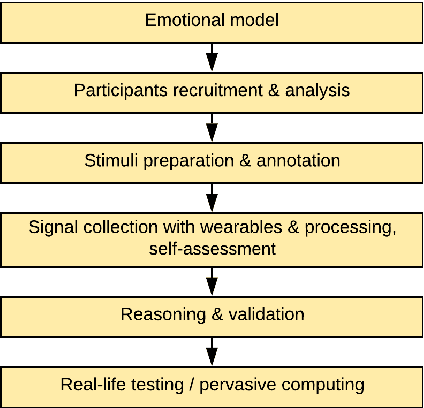
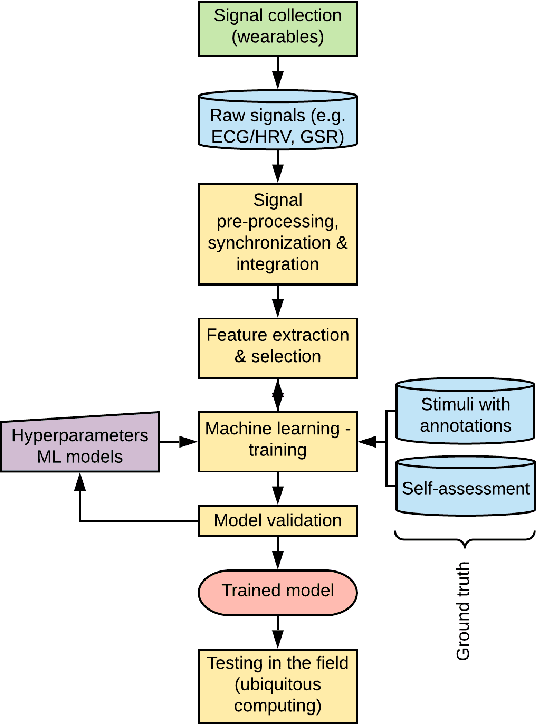

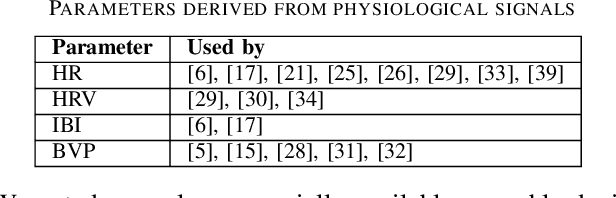
Abstract:Wearables like smartwatches or wrist bands equipped with pervasive sensors enable us to monitor our physiological signals. In this study, we address the question whether they can help us to recognize our emotions in our everyday life for ubiquitous computing. Using the systematic literature review, we identified crucial research steps and discussed the main limitations and problems in the domain.
An "outside the box" solution for imbalanced data classification
Nov 16, 2019



Abstract:A common problem of the real-world data sets is the class imbalance, which can significantly affect the classification abilities of classifiers. Numerous methods have been proposed to cope with this problem; however, even state-of-the-art methods offer a limited improvement (if any) for data sets with critically under-represented minority classes. For such problematic cases, an "outside the box" solution is required. Therefore, we propose a novel technique, called enrichment, which uses the information (observations) from the external data set(s). We present three approaches to implement enrichment technique: (1) selecting observations randomly, (2) iteratively choosing observations that improve the classification result, (3) adding observations that help the classifier to determine the border between classes better. We then thoroughly analyze developed solutions on ten real-world data sets to experimentally validate their usefulness. On average, our best approach improves the classification quality by 27\%, and in the best case, by outstanding 66\%. We also compare our technique with the universally applicable state-of-the-art methods. We find that our technique surpasses the existing methods performing, on average, 21\% better. The advantage is especially noticeable for the smallest data sets, for which existing methods failed, while our solutions achieved the best results. Additionally, our technique applies to both the multi-class and binary classification tasks. It can also be combined with other techniques dealing with the class imbalance problem.
Using Machine Learning to Predict the Evolution of Physics Research
Oct 29, 2018

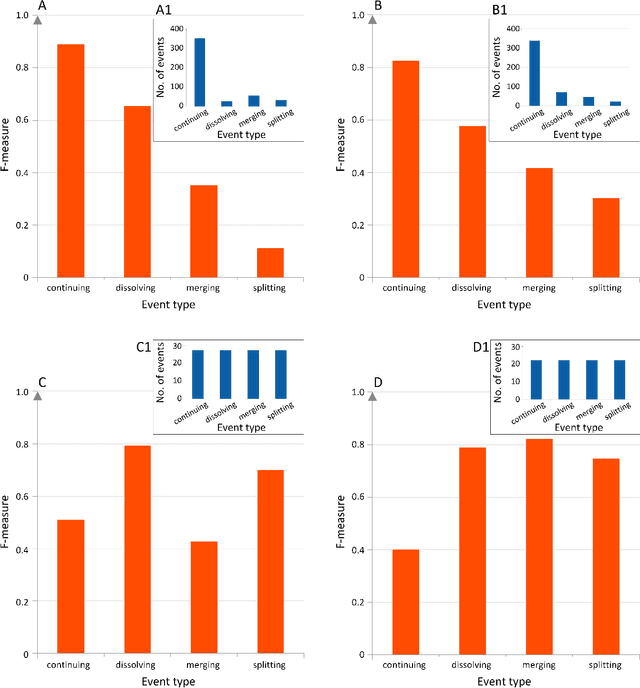
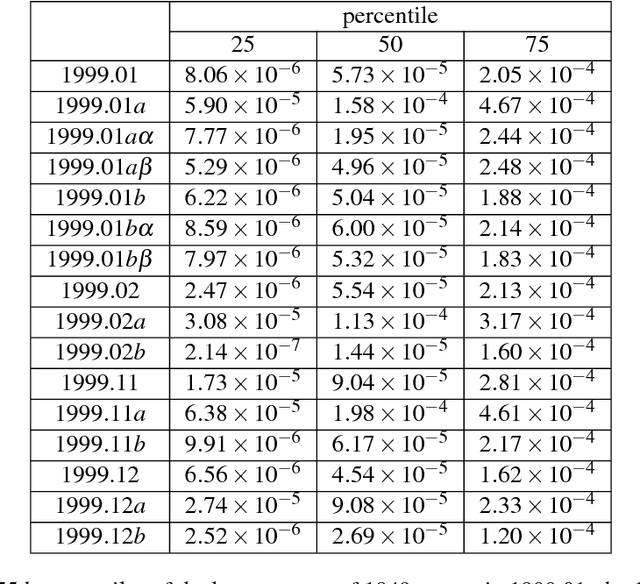
Abstract:The advancement of science as outlined by Popper and Kuhn is largely qualitative, but with bibliometric data it is possible and desirable to develop a quantitative picture of scientific progress. Furthermore it is also important to allocate finite resources to research topics that have growth potential, to accelerate the process from scientific breakthroughs to technological innovations. In this paper, we address this problem of quantitative knowledge evolution by analysing the APS publication data set from 1981 to 2010. We build the bibliographic coupling and co-citation networks, use the Louvain method to detect topical clusters (TCs) in each year, measure the similarity of TCs in consecutive years, and visualize the results as alluvial diagrams. Having the predictive features describing a given TC and its known evolution in the next year, we can train a machine learning model to predict future changes of TCs, i.e., their continuing, dissolving, merging and splitting. We found the number of papers from certain journals, the degree, closeness, and betweenness to be the most predictive features. Additionally, betweenness increases significantly for merging events, and decreases significantly for splitting events. Our results represent a first step from a descriptive understanding of the Science of Science (SciSci), towards one that is ultimately prescriptive.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge