Shixu Lin
Streamlining Social Media Information Retrieval for Public Health Research with Deep Learning
Jun 28, 2023Abstract:The utilization of social media in epidemic surveillance has been well established. Nonetheless, bias is often introduced when pre-defined lexicons are used to retrieve relevant corpus. This study introduces a framework aimed at curating extensive dictionaries of medical colloquialisms and Unified Medical Language System (UMLS) concepts. The framework comprises three modules: a BERT-based Named Entity Recognition (NER) model that identifies medical entities from social media content, a deep-learning powered normalization module that standardizes the extracted entities, and a semi-supervised clustering module that assigns the most probable UMLS concept to each standardized entity. We applied this framework to COVID-19-related tweets from February 1, 2020, to April 30, 2022, generating a symptom dictionary (available at https://github.com/ningkko/UMLS_colloquialism/) composed of 9,249 standardized entities mapped to 876 UMLS concepts and 38,175 colloquial expressions. This framework demonstrates encouraging potential in addressing the constraints of keyword matching information retrieval in social media-based public health research.
Exploring Social Media for Early Detection of Depression in COVID-19 Patients
Feb 23, 2023



Abstract:The COVID-19 pandemic has caused substantial damage to global health. Even though three years have passed, the world continues to struggle with the virus. Concerns are growing about the impact of COVID-19 on the mental health of infected individuals, who are more likely to experience depression, which can have long-lasting consequences for both the affected individuals and the world. Detection and intervention at an early stage can reduce the risk of depression in COVID-19 patients. In this paper, we investigated the relationship between COVID-19 infection and depression through social media analysis. Firstly, we managed a dataset of COVID-19 patients that contains information about their social media activity both before and after infection. Secondly,We conducted an extensive analysis of this dataset to investigate the characteristic of COVID-19 patients with a higher risk of depression. Thirdly, we proposed a deep neural network for early prediction of depression risk. This model considers daily mood swings as a psychiatric signal and incorporates textual and emotional characteristics via knowledge distillation. Experimental results demonstrate that our proposed framework outperforms baselines in detecting depression risk, with an AUROC of 0.9317 and an AUPRC of 0.8116. Our model has the potential to enable public health organizations to initiate prompt intervention with high-risk patients
Using Twitter Data to Understand Public Perceptions of Approved versus Off-label Use for COVID-19-related Medications
Jun 29, 2022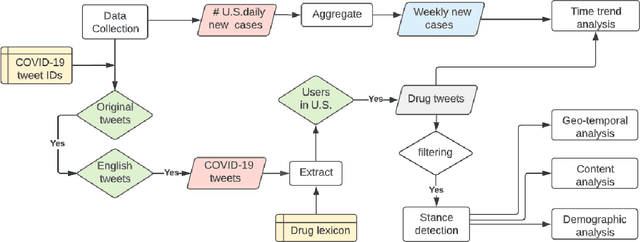

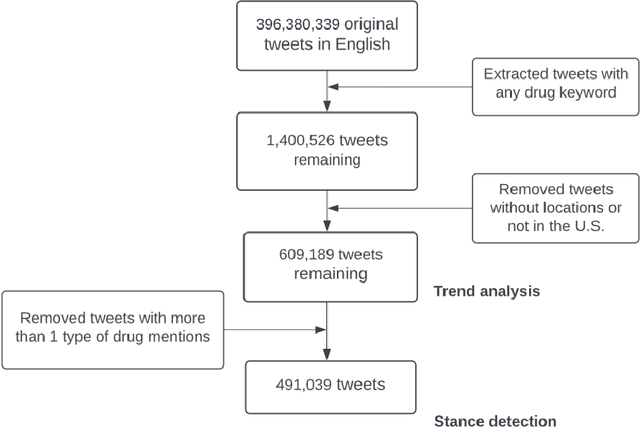
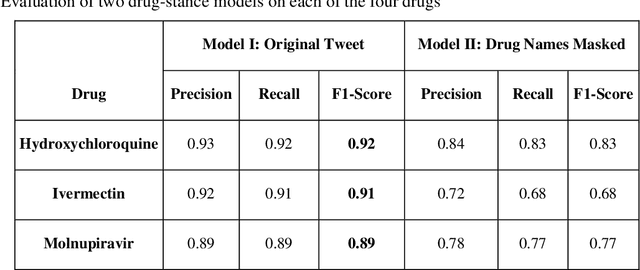
Abstract:Understanding public discourse on emergency use of unproven therapeutics is essential to monitor safe use and combat misinformation. We developed a natural language processing (NLP)-based pipeline to understand public perceptions of and stances on COVID-19-related drugs on Twitter across time. This retrospective study included 609,189 US-based tweets between January 29th, 2020 and November 30th, 2021 on four drugs that gained wide public attention during the COVID-19 pandemic: 1) Hydroxychloroquine and Ivermectin, drug therapies with anecdotal evidence; and 2) Molnupiravir and Remdesivir, FDA-approved treatment options for eligible patients. Time-trend analysis was used to understand the popularity and related events. Content and demographic analyses were conducted to explore potential rationales of people's stances on each drug. Time-trend analysis revealed that Hydroxychloroquine and Ivermectin received much more discussion than Molnupiravir and Remdesivir, particularly during COVID-19 surges. Hydroxychloroquine and Ivermectin were highly politicized, related to conspiracy theories, hearsay, celebrity effects, etc. The distribution of stance between the two major US political parties was significantly different (p<0.001); Republicans were much more likely to support Hydroxychloroquine (+55%) and Ivermectin (+30%) than Democrats. People with healthcare backgrounds tended to oppose Hydroxychloroquine (+7%) more than the general population; in contrast, the general population was more likely to support Ivermectin (+14%). We make all the data, code, and models available at https://github.com/ningkko/COVID-drug.
* This is a preliminary version. For full paper please refer to JAMIA
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge