Seyed Mohammad Jafar Jalali
Supervised Class-pairwise NMF for Data Representation and Classification
Sep 28, 2022



Abstract:Various Non-negative Matrix factorization (NMF) based methods add new terms to the cost function to adapt the model to specific tasks, such as clustering, or to preserve some structural properties in the reduced space (e.g., local invariance). The added term is mainly weighted by a hyper-parameter to control the balance of the overall formula to guide the optimization process towards the objective. The result is a parameterized NMF method. However, NMF method adopts unsupervised approaches to estimate the factorizing matrices. Thus, the ability to perform prediction (e.g. classification) using the new obtained features is not guaranteed. The objective of this work is to design an evolutionary framework to learn the hyper-parameter of the parameterized NMF and estimate the factorizing matrices in a supervised way to be more suitable for classification problems. Moreover, we claim that applying NMF-based algorithms separately to different class-pairs instead of applying it once to the whole dataset improves the effectiveness of the matrix factorization process. This results in training multiple parameterized NMF algorithms with different balancing parameter values. A cross-validation combination learning framework is adopted and a Genetic Algorithm is used to identify the optimal set of hyper-parameter values. The experiments we conducted on both real and synthetic datasets demonstrated the effectiveness of the proposed approach.
SpinalNet: Deep Neural Network with Gradual Input
Jul 07, 2020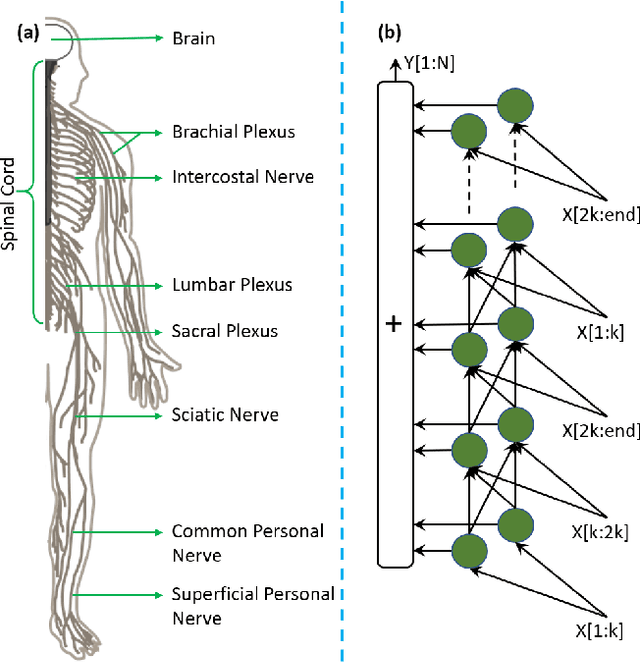
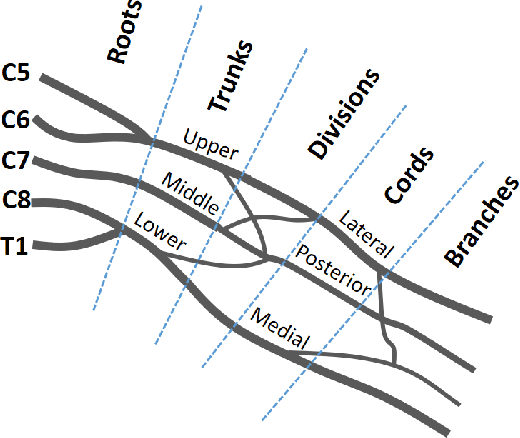
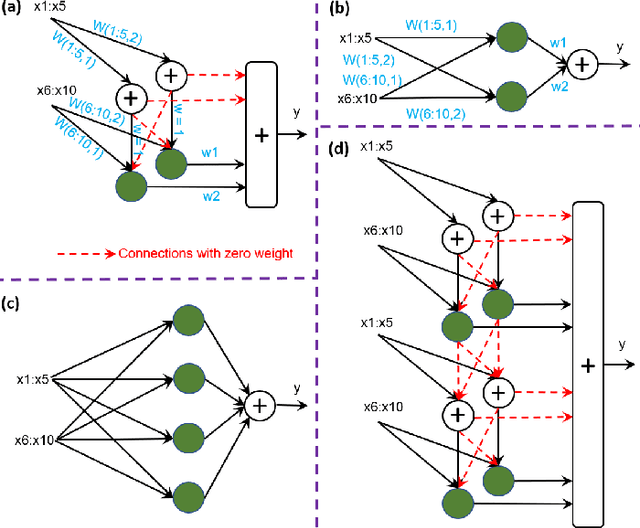
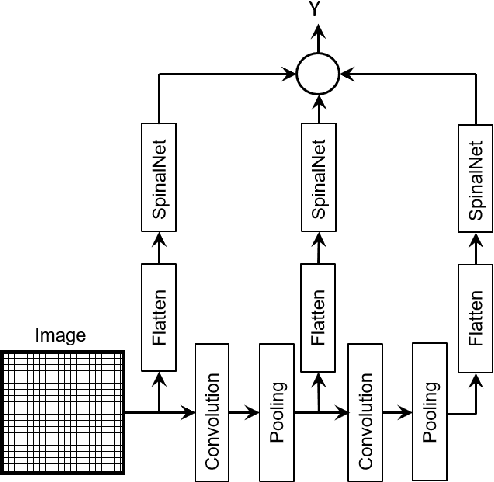
Abstract:Deep neural networks (DNNs) have achieved the state of the art performance in numerous fields. However, DNNs need high computation times, and people always expect better performance with lower computation. Therefore, we study the human somatosensory system and design a neural network (SpinalNet) to achieve higher accuracy with lower computation time. This paper aims to present the SpinalNet. Hidden layers of the proposed SpinalNet consist of three parts: 1) Input row, 2) Intermediate row, and 3) output row. The intermediate row of the SpinalNet usually contains a small number of neurons. Input segmentation enables each hidden layer to receive a part of the input and outputs of the previous layer. Therefore, the number of incoming weights in a hidden layer is significantly lower than traditional DNNs. As the network directly contributes to outputs in each layer, the vanishing gradient problem of DNN does not exist. We integrate the SpinalNet as the fully-connected layer of the convolutional neural network (CNN), residual neural network (ResNet), and Dense Convolutional Network (DenseNet), Visual Geometry Group (VGG) network. We observe a significant error reduction with lower computation in most situations. We have received state-of-the-art performance for the QMNIST, Kuzushiji-MNIST, and EMNIST(digits) datasets. Scripts of the proposed SpinalNet is available at the following link: https://github.com/dipuk0506/SpinalNet
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge