Seppo Virtanen
Local Language Models for Context-Aware Adaptive Anonymization of Sensitive Text
Jan 21, 2026Abstract:Qualitative research often contains personal, contextual, and organizational details that pose privacy risks if not handled appropriately. Manual anonymization is time-consuming, inconsistent, and frequently omits critical identifiers. Existing automated tools tend to rely on pattern matching or fixed rules, which fail to capture context and may alter the meaning of the data. This study uses local LLMs to build a reliable, repeatable, and context-aware anonymization process for detecting and anonymizing sensitive data in qualitative transcripts. We introduce a Structured Framework for Adaptive Anonymizer (SFAA) that includes three steps: detection, classification, and adaptive anonymization. The SFAA incorporates four anonymization strategies: rule-based substitution, context-aware rewriting, generalization, and suppression. These strategies are applied based on the identifier type and the risk level. The identifiers handled by the SFAA are guided by major international privacy and research ethics standards, including the GDPR, HIPAA, and OECD guidelines. This study followed a dual-method evaluation that combined manual and LLM-assisted processing. Two case studies were used to support the evaluation. The first includes 82 face-to-face interviews on gamification in organizations. The second involves 93 machine-led interviews using an AI-powered interviewer to test LLM awareness and workplace privacy. Two local models, LLaMA and Phi were used to evaluate the performance of the proposed framework. The results indicate that the LLMs found more sensitive data than a human reviewer. Phi outperformed LLaMA in finding sensitive data, but made slightly more errors. Phi was able to find over 91% of the sensitive data and 94.8% kept the same sentiment as the original text, which means it was very accurate, hence, it does not affect the analysis of the qualitative data.
A Generative Shape Compositional Framework: Towards Representative Populations of Virtual Heart Chimaeras
Oct 04, 2022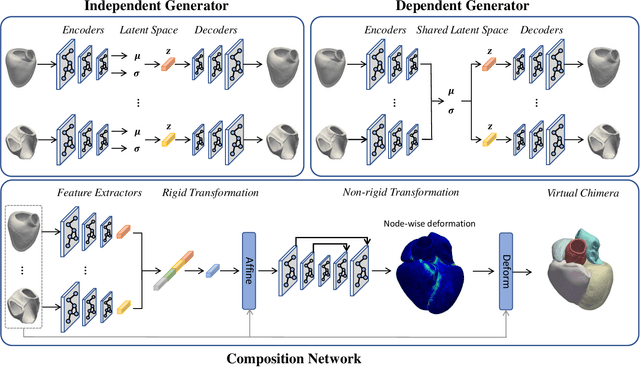
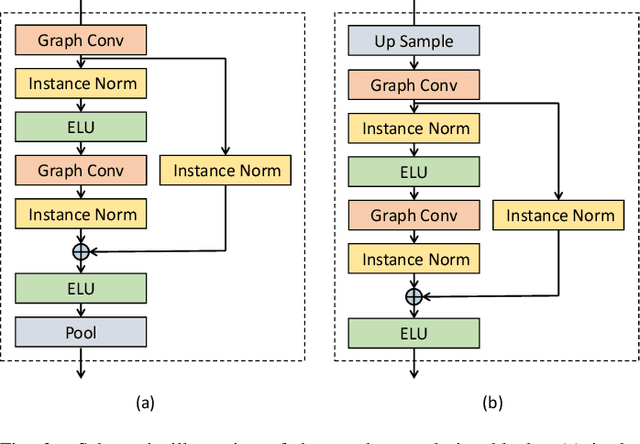
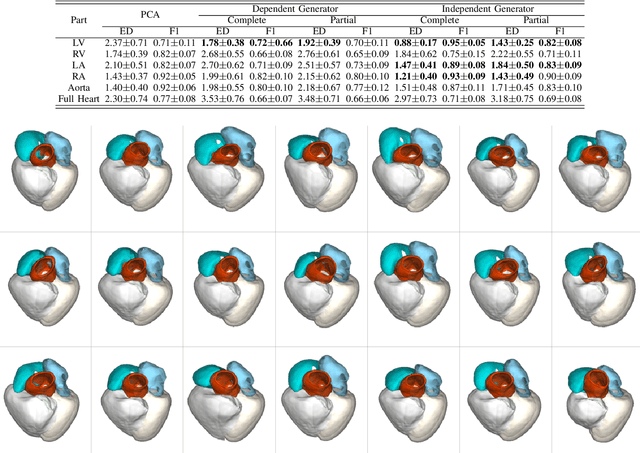
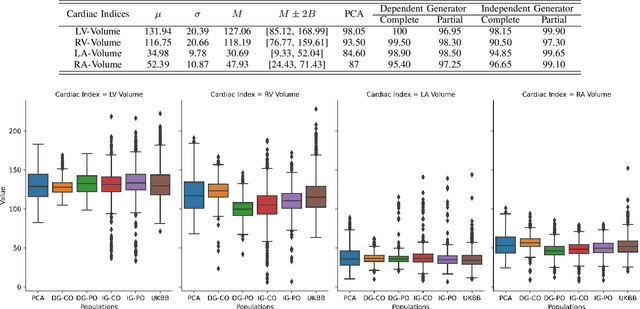
Abstract:Generating virtual populations of anatomy that capture sufficient variability while remaining plausible is essential for conducting in-silico trials of medical devices. However, not all anatomical shapes of interest are always available for each individual in a population. Hence, missing/partially-overlapping anatomical information is often available across individuals in a population. We introduce a generative shape model for complex anatomical structures, learnable from datasets of unpaired datasets. The proposed generative model can synthesise complete whole complex shape assemblies coined virtual chimaeras, as opposed to natural human chimaeras. We applied this framework to build virtual chimaeras from databases of whole-heart shape assemblies that each contribute samples for heart substructures. Specifically, we propose a generative shape compositional framework which comprises two components - a part-aware generative shape model which captures the variability in shape observed for each structure of interest in the training population; and a spatial composition network which assembles/composes the structures synthesised by the former into multi-part shape assemblies (viz. virtual chimaeras). We also propose a novel self supervised learning scheme that enables the spatial composition network to be trained with partially overlapping data and weak labels. We trained and validated our approach using shapes of cardiac structures derived from cardiac magnetic resonance images available in the UK Biobank. Our approach significantly outperforms a PCA-based shape model (trained with complete data) in terms of generalisability and specificity. This demonstrates the superiority of the proposed approach as the synthesised cardiac virtual populations are more plausible and capture a greater degree of variability in shape than those generated by the PCA-based shape model.
Visualizations Relevant to The User By Multi-View Latent Variable Factorization
Jan 25, 2016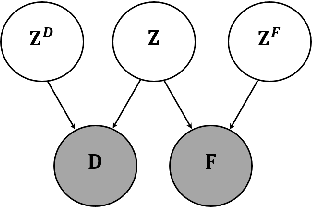
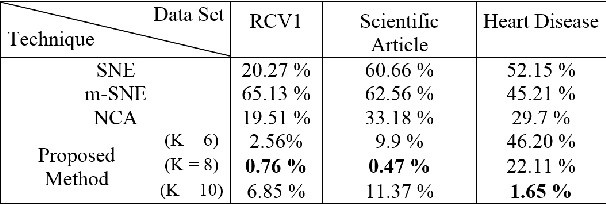
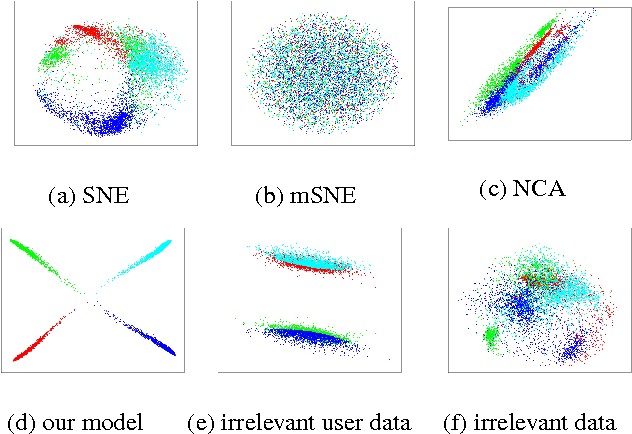
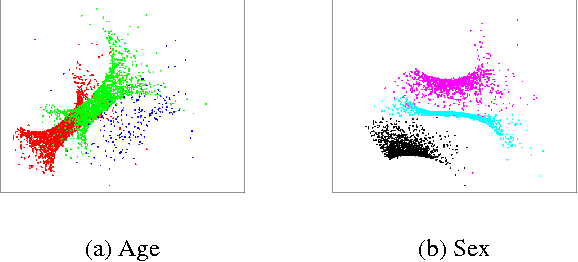
Abstract:A main goal of data visualization is to find, from among all the available alternatives, mappings to the 2D/3D display which are relevant to the user. Assuming user interaction data, or other auxiliary data about the items or their relationships, the goal is to identify which aspects in the primary data support the user\'s input and, equally importantly, which aspects of the user\'s potentially noisy input have support in the primary data. For solving the problem, we introduce a multi-view embedding in which a latent factorization identifies which aspects in the two data views (primary data and user data) are related and which are specific to only one of them. The factorization is a generative model in which the display is parameterized as a part of the factorization and the other factors explain away the aspects not expressible in a two-dimensional display. Functioning of the model is demonstrated on several data sets.
Group Factor Analysis
Dec 02, 2014



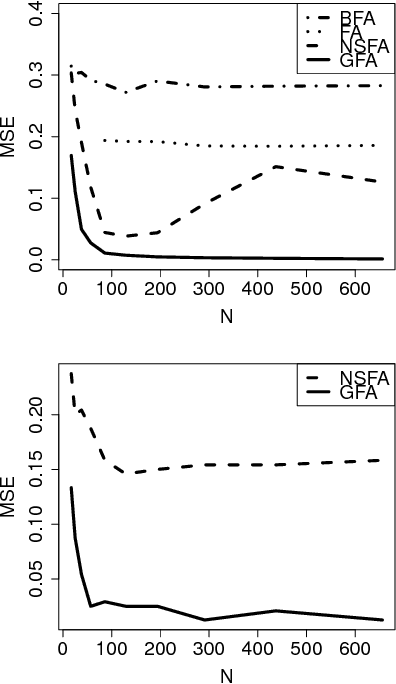
Abstract:Factor analysis provides linear factors that describe relationships between individual variables of a data set. We extend this classical formulation into linear factors that describe relationships between groups of variables, where each group represents either a set of related variables or a data set. The model also naturally extends canonical correlation analysis to more than two sets, in a way that is more flexible than previous extensions. Our solution is formulated as variational inference of a latent variable model with structural sparsity, and it consists of two hierarchical levels: The higher level models the relationships between the groups, whereas the lower models the observed variables given the higher level. We show that the resulting solution solves the group factor analysis problem accurately, outperforming alternative factor analysis based solutions as well as more straightforward implementations of group factor analysis. The method is demonstrated on two life science data sets, one on brain activation and the other on systems biology, illustrating its applicability to the analysis of different types of high-dimensional data sources.
Identification of structural features in chemicals associated with cancer drug response: A systematic data-driven analysis
Apr 29, 2014
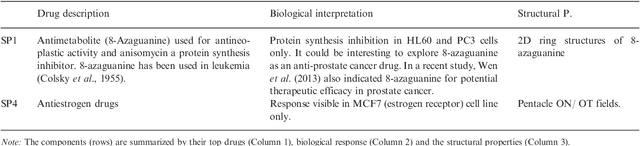


Abstract:Motivation: Analysis of relationships of drug structure to biological response is key to understanding off-target and unexpected drug effects, and for developing hypotheses on how to tailor drug thera-pies. New methods are required for integrated analyses of a large number of chemical features of drugs against the corresponding genome-wide responses of multiple cell models. Results: In this paper, we present the first comprehensive multi-set analysis on how the chemical structure of drugs impacts on ge-nome-wide gene expression across several cancer cell lines (CMap database). The task is formulated as searching for drug response components across multiple cancers to reveal shared effects of drugs and the chemical features that may be responsible. The com-ponents can be computed with an extension of a very recent ap-proach called Group Factor Analysis (GFA). We identify 11 compo-nents that link the structural descriptors of drugs with specific gene expression responses observed in the three cell lines, and identify structural groups that may be responsible for the responses. Our method quantitatively outperforms the limited earlier studies on CMap and identifies both the previously reported associations and several interesting novel findings, by taking into account multiple cell lines and advanced 3D structural descriptors. The novel observations include: previously unknown similarities in the effects induced by 15-delta prostaglandin J2 and HSP90 inhibitors, which are linked to the 3D descriptors of the drugs; and the induction by simvastatin of leukemia-specific anti-inflammatory response, resem-bling the effects of corticosteroids.
Factorized Multi-Modal Topic Model
Oct 16, 2012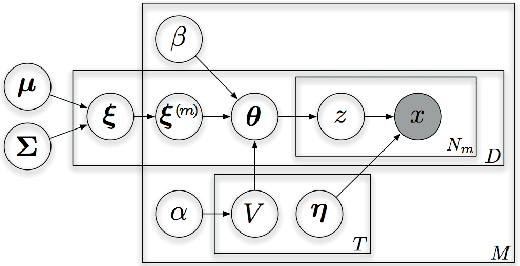



Abstract:Multi-modal data collections, such as corpora of paired images and text snippets, require analysis methods beyond single-view component and topic models. For continuous observations the current dominant approach is based on extensions of canonical correlation analysis, factorizing the variation into components shared by the different modalities and those private to each of them. For count data, multiple variants of topic models attempting to tie the modalities together have been presented. All of these, however, lack the ability to learn components private to one modality, and consequently will try to force dependencies even between minimally correlating modalities. In this work we combine the two approaches by presenting a novel HDP-based topic model that automatically learns both shared and private topics. The model is shown to be especially useful for querying the contents of one domain given samples of the other.
Bayesian exponential family projections for coupled data sources
Mar 15, 2012



Abstract:Exponential family extensions of principal component analysis (EPCA) have received a considerable amount of attention in recent years, demonstrating the growing need for basic modeling tools that do not assume the squared loss or Gaussian distribution. We extend the EPCA model toolbox by presenting the first exponential family multi-view learning methods of the partial least squares and canonical correlation analysis, based on a unified representation of EPCA as matrix factorization of the natural parameters of exponential family. The models are based on a new family of priors that are generally usable for all such factorizations. We also introduce new inference strategies, and demonstrate how the methods outperform earlier ones when the Gaussianity assumption does not hold.
Bayesian Group Factor Analysis
Oct 14, 2011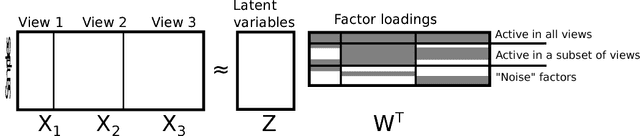
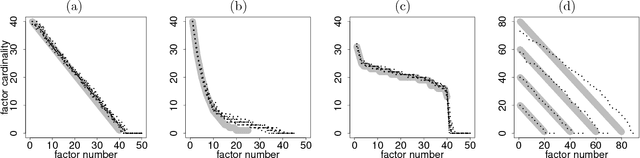
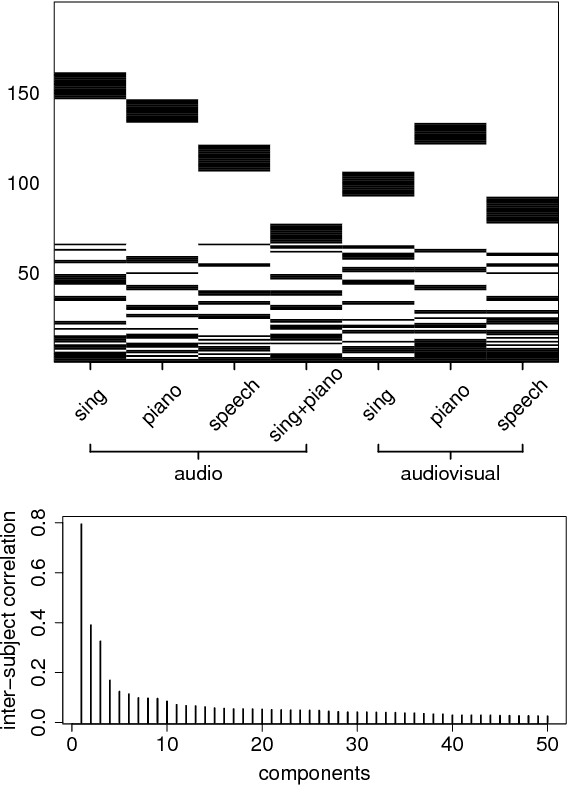
Abstract:We introduce a factor analysis model that summarizes the dependencies between observed variable groups, instead of dependencies between individual variables as standard factor analysis does. A group may correspond to one view of the same set of objects, one of many data sets tied by co-occurrence, or a set of alternative variables collected from statistics tables to measure one property of interest. We show that by assuming group-wise sparse factors, active in a subset of the sets, the variation can be decomposed into factors explaining relationships between the sets and factors explaining away set-specific variation. We formulate the assumptions in a Bayesian model which provides the factors, and apply the model to two data analysis tasks, in neuroimaging and chemical systems biology.
* 9 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge