Suleiman A Khan
Convex Factorization Machine for Regression
Aug 10, 2016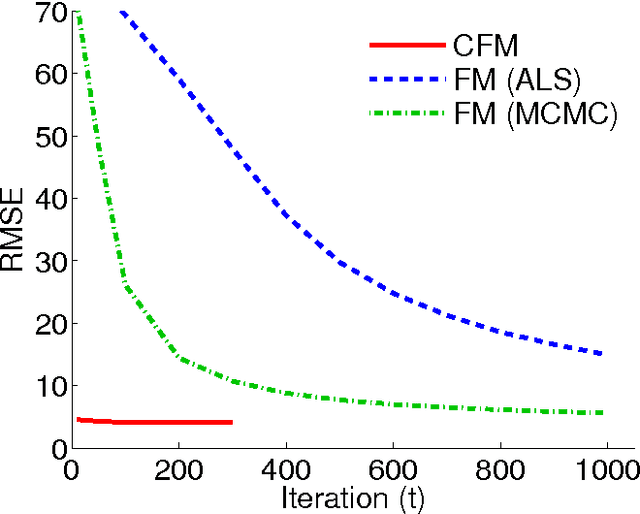
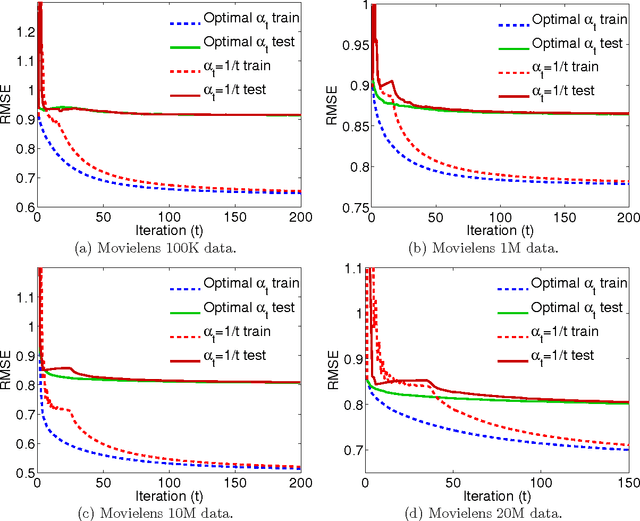
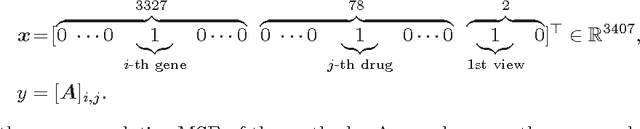
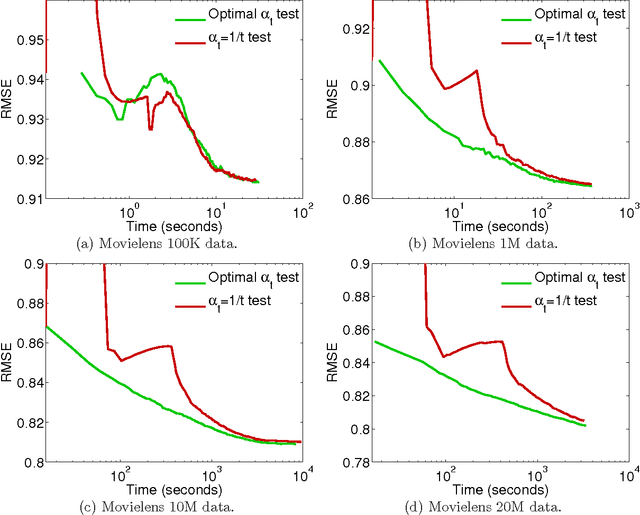
Abstract:We propose the convex factorization machine (CFM), which is a convex variant of the widely used Factorization Machines (FMs). Specifically, we employ a linear+quadratic model and regularize the linear term with the $\ell_2$-regularizer and the quadratic term with the trace norm regularizer. Then, we formulate the CFM optimization as a semidefinite programming problem and propose an efficient optimization procedure with Hazan's algorithm. A key advantage of CFM over existing FMs is that it can find a globally optimal solution, while FMs may get a poor locally optimal solution since the objective function of FMs is non-convex. In addition, the proposed algorithm is simple yet effective and can be implemented easily. Finally, CFM is a general factorization method and can also be used for other factorization problems including including multi-view matrix factorization and tensor completion problems. Through synthetic and movielens datasets, we first show that the proposed CFM achieves results competitive to FMs. Furthermore, in a toxicogenomics prediction task, we show that CFM outperforms a state-of-the-art tensor factorization method.
Identification of structural features in chemicals associated with cancer drug response: A systematic data-driven analysis
Apr 29, 2014
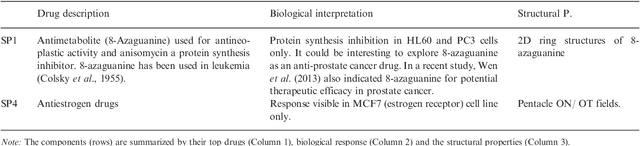


Abstract:Motivation: Analysis of relationships of drug structure to biological response is key to understanding off-target and unexpected drug effects, and for developing hypotheses on how to tailor drug thera-pies. New methods are required for integrated analyses of a large number of chemical features of drugs against the corresponding genome-wide responses of multiple cell models. Results: In this paper, we present the first comprehensive multi-set analysis on how the chemical structure of drugs impacts on ge-nome-wide gene expression across several cancer cell lines (CMap database). The task is formulated as searching for drug response components across multiple cancers to reveal shared effects of drugs and the chemical features that may be responsible. The com-ponents can be computed with an extension of a very recent ap-proach called Group Factor Analysis (GFA). We identify 11 compo-nents that link the structural descriptors of drugs with specific gene expression responses observed in the three cell lines, and identify structural groups that may be responsible for the responses. Our method quantitatively outperforms the limited earlier studies on CMap and identifies both the previously reported associations and several interesting novel findings, by taking into account multiple cell lines and advanced 3D structural descriptors. The novel observations include: previously unknown similarities in the effects induced by 15-delta prostaglandin J2 and HSP90 inhibitors, which are linked to the 3D descriptors of the drugs; and the induction by simvastatin of leukemia-specific anti-inflammatory response, resem-bling the effects of corticosteroids.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge