Sailun Xu
Image Style Transfer and Content-Style Disentanglement
Nov 25, 2021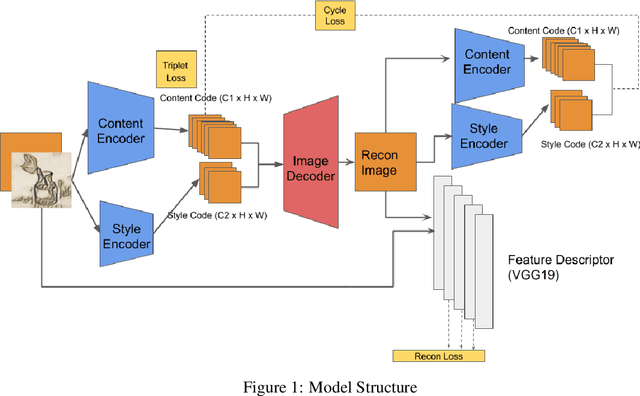
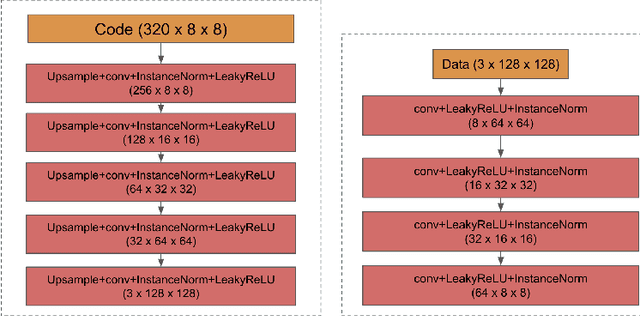
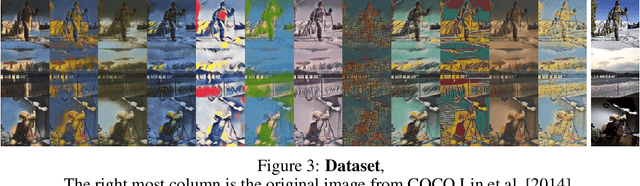
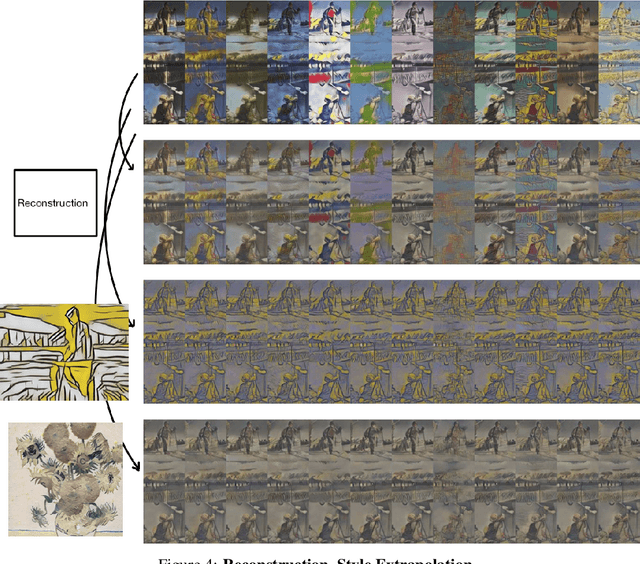
Abstract:We propose a way of learning disentangled content-style representation of image, allowing us to extrapolate images to any style as well as interpolate between any pair of styles. By augmenting data set in a supervised setting and imposing triplet loss, we ensure the separation of information encoded by content and style representation. We also make use of cycle-consistency loss to guarantee that images could be reconstructed faithfully by their representation.
Video Content Swapping Using GAN
Nov 21, 2021



Abstract:Video generation is an interesting problem in computer vision. It is quite popular for data augmentation, special effect in move, AR/VR and so on. With the advances of deep learning, many deep generative models have been proposed to solve this task. These deep generative models provide away to utilize all the unlabeled images and videos online, since it can learn deep feature representations with unsupervised manner. These models can also generate different kinds of images, which have great value for visual application. However generating a video would be much more challenging since we need to model not only the appearances of objects in the video but also their temporal motion. In this work, we will break down any frame in the video into content and pose. We first extract the pose information from a video using a pre-trained human pose detection and use a generative model to synthesize the video based on the content code and pose code.
ChemBO: Bayesian Optimization of Small Organic Molecules with Synthesizable Recommendations
Aug 05, 2019



Abstract:We describe ChemBO, a Bayesian Optimization framework for generating and optimizing organic molecules for desired molecular properties. This framework is useful in applications such as drug discovery, where an algorithm recommends new candidate molecules; these molecules first need to be synthesized and then tested for drug-like properties. The algorithm uses the results of past tests to recommend new ones so as to find good molecules efficiently. Most existing data-driven methods for this problem do not account for sample efficiency and/or fail to enforce realistic constraints on synthesizability. In this work, we explore existing kernels for molecules in the literature as well as propose a novel kernel which views a molecule as a graph. In ChemBO, we implement these kernels in a Gaussian process model. Then we explore the chemical space by traversing possible paths of molecular synthesis. Consequently, our approach provides a proposal synthesis path every time it recommends a new molecule to test, a crucial advantage when compared to existing methods. In our experiments, we demonstrate the efficacy of the proposed approach on several molecular optimization problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge