Tingfung Lau
Quantifying and Understanding Adversarial Examples in Discrete Input Spaces
Dec 12, 2021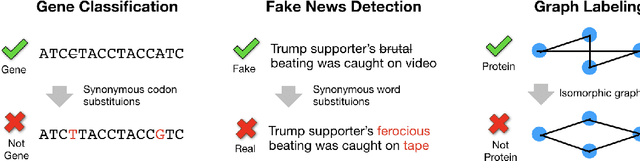
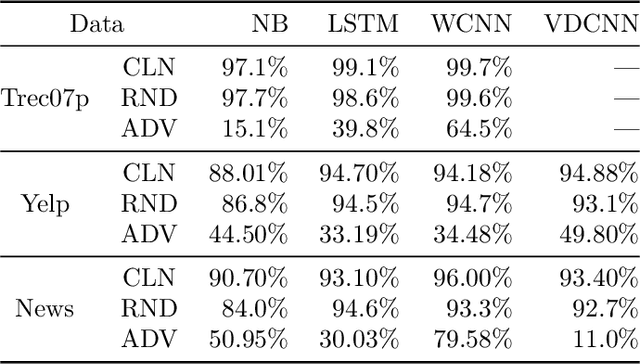
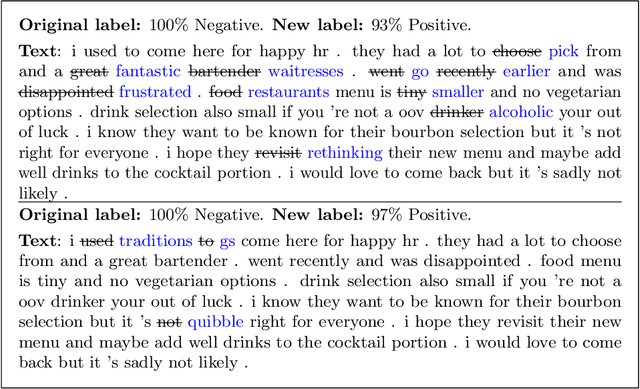

Abstract:Modern classification algorithms are susceptible to adversarial examples--perturbations to inputs that cause the algorithm to produce undesirable behavior. In this work, we seek to understand and extend adversarial examples across domains in which inputs are discrete, particularly across new domains, such as computational biology. As a step towards this goal, we formalize a notion of synonymous adversarial examples that applies in any discrete setting and describe a simple domain-agnostic algorithm to construct such examples. We apply this algorithm across multiple domains--including sentiment analysis and DNA sequence classification--and find that it consistently uncovers adversarial examples. We seek to understand their prevalence theoretically and we attribute their existence to spurious token correlations, a statistical phenomenon that is specific to discrete spaces. Our work is a step towards a domain-agnostic treatment of discrete adversarial examples analogous to that of continuous inputs.
Video Content Swapping Using GAN
Nov 21, 2021



Abstract:Video generation is an interesting problem in computer vision. It is quite popular for data augmentation, special effect in move, AR/VR and so on. With the advances of deep learning, many deep generative models have been proposed to solve this task. These deep generative models provide away to utilize all the unlabeled images and videos online, since it can learn deep feature representations with unsupervised manner. These models can also generate different kinds of images, which have great value for visual application. However generating a video would be much more challenging since we need to model not only the appearances of objects in the video but also their temporal motion. In this work, we will break down any frame in the video into content and pose. We first extract the pose information from a video using a pre-trained human pose detection and use a generative model to synthesize the video based on the content code and pose code.
Embryo staging with weakly-supervised region selection and dynamically-decoded predictions
Apr 09, 2019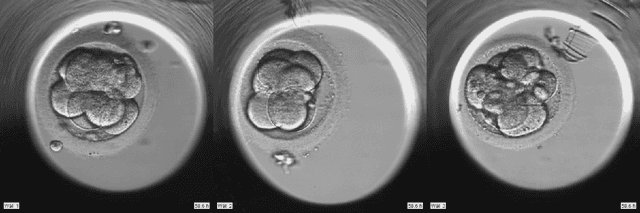
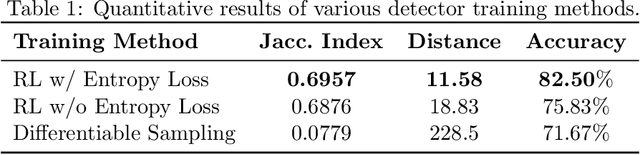
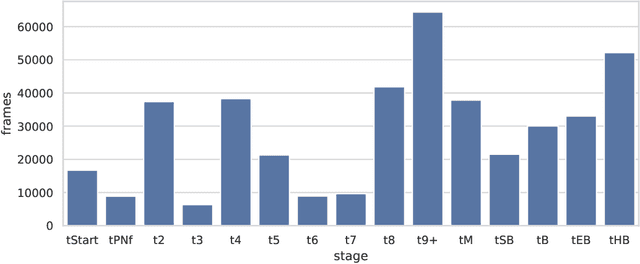
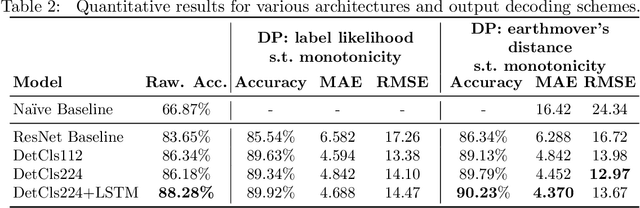
Abstract:To optimize clinical outcomes, fertility clinics must strategically select which embryos to transfer. Common selection heuristics are formulas expressed in terms of the durations required to reach various developmental milestones, quantities historically annotated manually by experienced embryologists based on time-lapse EmbryoScope videos. We propose a new method for automatic embryo staging that exploits several sources of structure in this time-lapse data. First, noting that in each image the embryo occupies a small subregion, we jointly train a region proposal network with the downstream classifier to isolate the embryo. Notably, because we lack ground-truth bounding boxes, our we weakly supervise the region proposal network optimizing its parameters via reinforcement learning to improve the downstream classifier's loss. Moreover, noting that embryos reaching the blastocyst stage progress monotonically through earlier stages, we develop a dynamic-programming-based decoder that post-processes our predictions to select the most likely monotonic sequence of developmental stages. Our methods outperform vanilla residual networks and rival the best numbers in contemporary papers, as measured by both per-frame accuracy and transition prediction error, despite operating on smaller data than many.
Unsupervised 3D End-to-End Medical Image Registration with Volume Tweening Network
Feb 13, 2019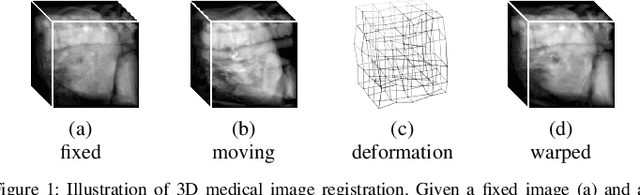
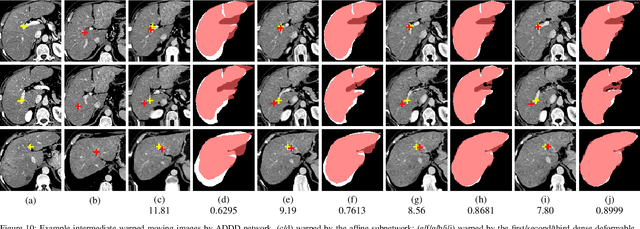
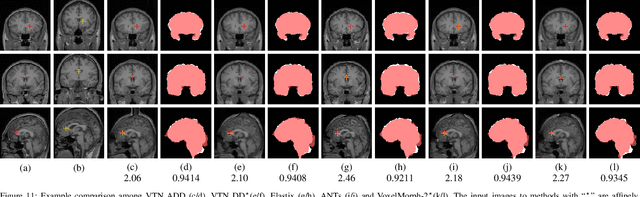
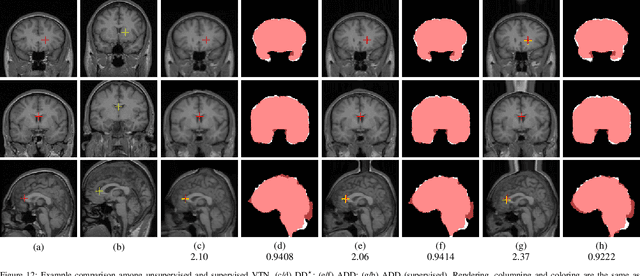
Abstract:3D medical image registration is of great clinical importance. However, supervised learning methods require a large amount of accurately annotated corresponding control points (or morphing). The ground truth for 3D medical images is very difficult to obtain. Unsupervised learning methods ease the burden of manual annotation by exploiting unlabeled data without supervision. In this paper, we propose a new unsupervised learning method using convolutional neural networks under an end-to-end framework, Volume Tweening Network (VTN), to register 3D medical images. Three technical components ameliorate our unsupervised learning system for 3D end-to-end medical image registration: (1) We cascade the registration subnetworks; (2) We integrate affine registration into our network; and (3) We incorporate an additional invertibility loss into the training process. Experimental results demonstrate that our algorithm is 880x faster (or 3.3x faster without GPU acceleration) than traditional optimization-based methods and achieves state-of-the-art performance in medical image registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge