Ryota Maeda
Dense Dispersed Structured Light for Hyperspectral 3D Imaging of Dynamic Scenes
Dec 02, 2024


Abstract:Hyperspectral 3D imaging captures both depth maps and hyperspectral images, enabling comprehensive geometric and material analysis. Recent methods achieve high spectral and depth accuracy; however, they require long acquisition times often over several minutes or rely on large, expensive systems, restricting their use to static scenes. We present Dense Dispersed Structured Light (DDSL), an accurate hyperspectral 3D imaging method for dynamic scenes that utilizes stereo RGB cameras and an RGB projector equipped with an affordable diffraction grating film. We design spectrally multiplexed DDSL patterns that significantly reduce the number of required projector patterns, thereby accelerating acquisition speed. Additionally, we formulate an image formation model and a reconstruction method to estimate a hyperspectral image and depth map from captured stereo images. As the first practical and accurate hyperspectral 3D imaging method for dynamic scenes, we experimentally demonstrate that DDSL achieves a spectral resolution of 15.5 nm full width at half maximum (FWHM), a depth error of 4 mm, and a frame rate of 6.6 fps.
Event Ellipsometer: Event-based Mueller-Matrix Video Imaging
Nov 26, 2024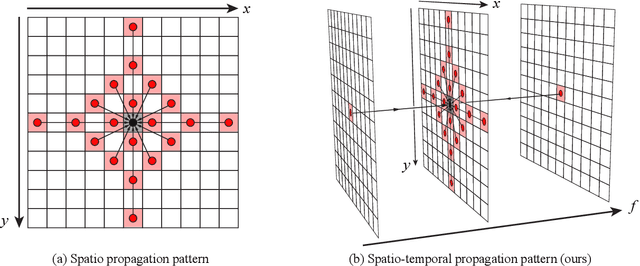
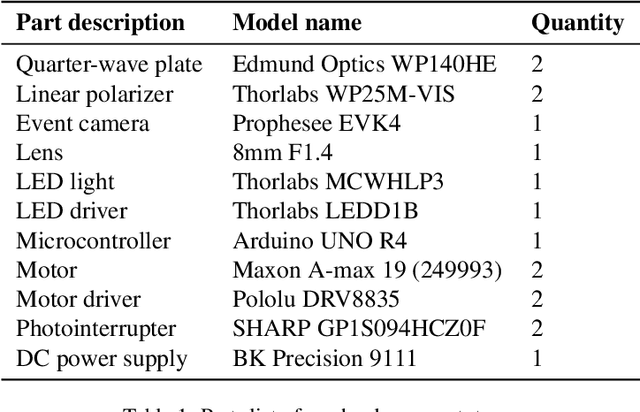
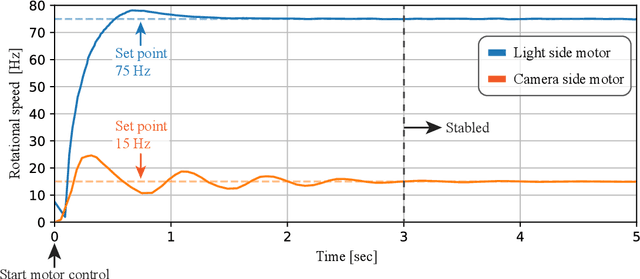
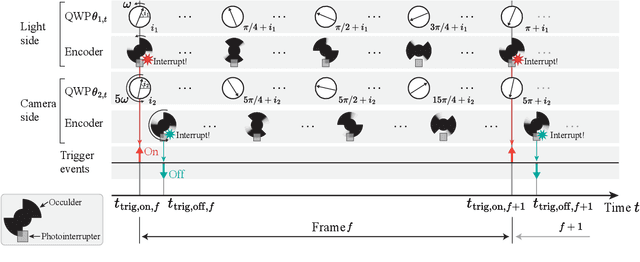
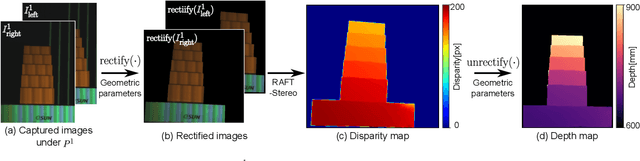
Abstract:Light-matter interactions modify both the intensity and polarization state of light. Changes in polarization, represented by a Mueller matrix, encode detailed scene information. Existing optical ellipsometers capture Mueller-matrix images; however, they are often limited to capturing static scenes due to long acquisition times. Here, we introduce Event Ellipsometer, a method for acquiring a Mueller-matrix video for dynamic scenes. Our imaging system employs fast-rotating quarter-wave plates (QWPs) in front of a light source and an event camera that asynchronously captures intensity changes induced by the rotating QWPs. We develop an ellipsometric-event image formation model, a calibration method, and an ellipsometric-event reconstruction method. We experimentally demonstrate that Event Ellipsometer enables Mueller-matrix video imaging at 30fps, extending ellipsometry to dynamic scenes.
A SARS-CoV-2 Interaction Dataset and VHH Sequence Corpus for Antibody Language Models
May 29, 2024



Abstract:Antibodies are crucial proteins produced by the immune system to eliminate harmful foreign substances and have become pivotal therapeutic agents for treating human diseases. To accelerate the discovery of antibody therapeutics, there is growing interest in constructing language models using antibody sequences. However, the applicability of pre-trained language models for antibody discovery has not been thoroughly evaluated due to the scarcity of labeled datasets. To overcome these limitations, we introduce AVIDa-SARS-CoV-2, a dataset featuring the antigen-variable domain of heavy chain of heavy chain antibody (VHH) interactions obtained from two alpacas immunized with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike proteins. AVIDa-SARS-CoV-2 includes binary labels indicating the binding or non-binding of diverse VHH sequences to 12 SARS-CoV-2 mutants, such as the Delta and Omicron variants. Furthermore, we release VHHCorpus-2M, a pre-training dataset for antibody language models, containing over two million VHH sequences. We report benchmark results for predicting SARS-CoV-2-VHH binding using VHHBERT pre-trained on VHHCorpus-2M and existing general protein and antibody-specific pre-trained language models. These results confirm that AVIDa-SARS-CoV-2 provides valuable benchmarks for evaluating the representation capabilities of antibody language models for binding prediction, thereby facilitating the development of AI-driven antibody discovery. The datasets are available at https://datasets.cognanous.com.
Polarimetric Light Transport Analysis for Specular Inter-reflection
Dec 07, 2023



Abstract:Polarization is well known for its ability to decompose diffuse and specular reflections. However, the existing decomposition methods only focus on direct reflection and overlook multiple reflections, especially specular inter-reflection. In this paper, we propose a novel decomposition method for handling specular inter-reflection of metal objects by using a unique polarimetric feature: the rotation direction of linear polarization. This rotation direction serves as a discriminative factor between direct and inter-reflection on specular surfaces. To decompose the reflectance components, we actively rotate the linear polarization of incident light and analyze the rotation direction of the reflected light. We evaluate our method using both synthetic and real data, demonstrating its effectiveness in decomposing specular inter-reflections of metal objects. Furthermore, we demonstrate that our method can be combined with other decomposition methods for a detailed analysis of light transport. As a practical application, we show its effectiveness in improving the accuracy of 3D measurement against strong specular inter-reflection.
AVIDa-hIL6: A Large-Scale VHH Dataset Produced from an Immunized Alpaca for Predicting Antigen-Antibody Interactions
Jun 06, 2023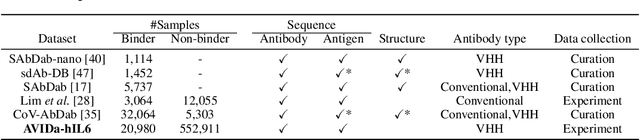
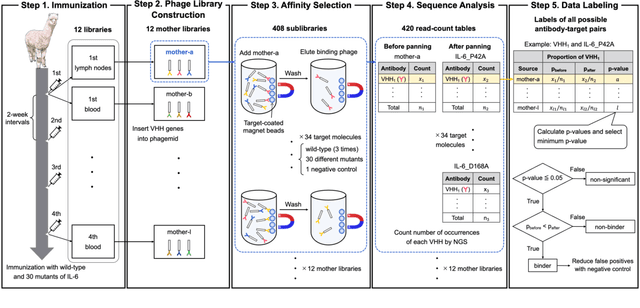
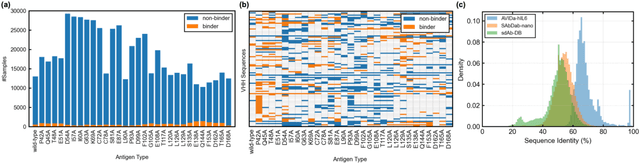

Abstract:Antibodies have become an important class of therapeutic agents to treat human diseases. To accelerate therapeutic antibody discovery, computational methods, especially machine learning, have attracted considerable interest for predicting specific interactions between antibody candidates and target antigens such as viruses and bacteria. However, the publicly available datasets in existing works have notable limitations, such as small sizes and the lack of non-binding samples and exact amino acid sequences. To overcome these limitations, we have developed AVIDa-hIL6, a large-scale dataset for predicting antigen-antibody interactions in the variable domain of heavy chain of heavy chain antibodies (VHHs), produced from an alpaca immunized with the human interleukin-6 (IL-6) protein, as antigens. By leveraging the simple structure of VHHs, which facilitates identification of full-length amino acid sequences by DNA sequencing technology, AVIDa-hIL6 contains 573,891 antigen-VHH pairs with amino acid sequences. All the antigen-VHH pairs have reliable labels for binding or non-binding, as generated by a novel labeling method. Furthermore, via introduction of artificial mutations, AVIDa-hIL6 contains 30 different mutants in addition to wild-type IL-6 protein. This characteristic provides opportunities to develop machine learning models for predicting changes in antibody binding by antigen mutations. We report experimental benchmark results on AVIDa-hIL6 by using neural network-based baseline models. The results indicate that the existing models have potential, but further research is needed to generalize them to predict effective antibodies against unknown mutants. The dataset is available at https://avida-hil6.cognanous.com.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge