Roshan Rao
ZPD Teaching Strategies for Deep Reinforcement Learning from Demonstrations
Oct 26, 2019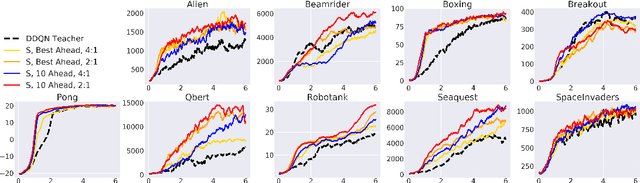
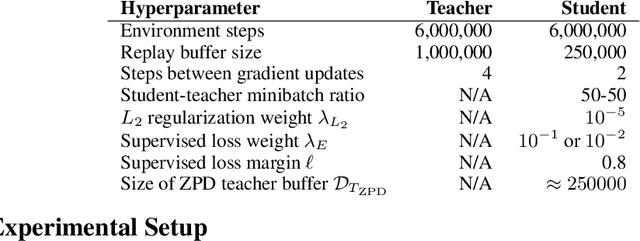
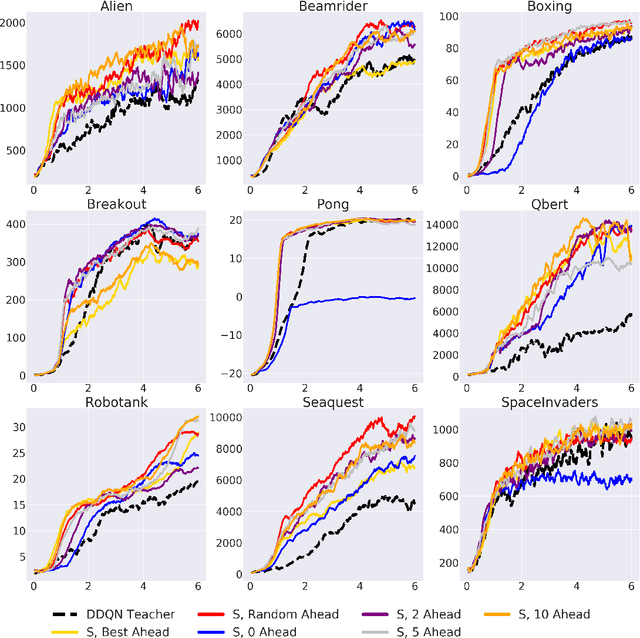
Abstract:Learning from demonstrations is a popular tool for accelerating and reducing the exploration requirements of reinforcement learning. When providing expert demonstrations to human students, we know that the demonstrations must fall within a particular range of difficulties called the "Zone of Proximal Development (ZPD)". If they are too easy the student learns nothing, but if they are too difficult the student is unable to follow along. This raises the question: Given a set of potential demonstrators, which among them is best suited for teaching any particular learner? Prior work, such as the popular Deep Q-learning from Demonstrations (DQfD) algorithm has generally focused on single demonstrators. In this work we consider the problem of choosing among multiple demonstrators of varying skill levels. Our results align with intuition from human learners: it is not always the best policy to draw demonstrations from the best performing demonstrator (in terms of reward). We show that careful selection of teaching strategies can result in sample efficiency gains in the learner's environment across nine Atari games
Evaluating Protein Transfer Learning with TAPE
Jun 19, 2019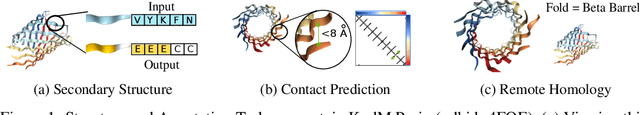
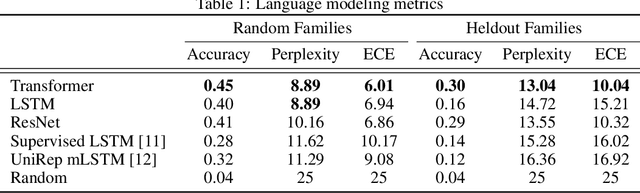
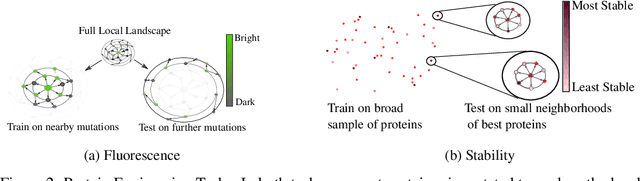
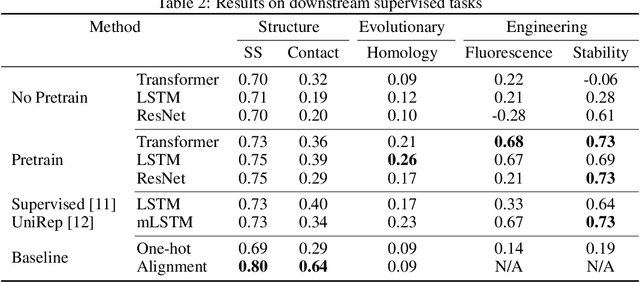
Abstract:Protein modeling is an increasingly popular area of machine learning research. Semi-supervised learning has emerged as an important paradigm in protein modeling due to the high cost of acquiring supervised protein labels, but the current literature is fragmented when it comes to datasets and standardized evaluation techniques. To facilitate progress in this field, we introduce the Tasks Assessing Protein Embeddings (TAPE), a set of five biologically relevant semi-supervised learning tasks spread across different domains of protein biology. We curate tasks into specific training, validation, and test splits to ensure that each task tests biologically relevant generalization that transfers to real-life scenarios. We benchmark a range of approaches to semi-supervised protein representation learning, which span recent work as well as canonical sequence learning techniques. We find that self-supervised pretraining is helpful for almost all models on all tasks, more than doubling performance in some cases. Despite this increase, in several cases features learned by self-supervised pretraining still lag behind features extracted by state-of-the-art non-neural techniques. This gap in performance suggests a huge opportunity for innovative architecture design and improved modeling paradigms that better capture the signal in biological sequences. TAPE will help the machine learning community focus effort on scientifically relevant problems. Toward this end, all data and code used to run these experiments are available at https://github.com/songlab-cal/tape.
t-SNE-CUDA: GPU-Accelerated t-SNE and its Applications to Modern Data
Jul 31, 2018
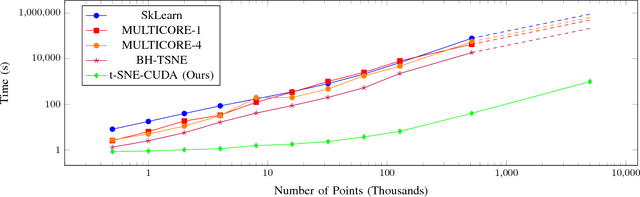
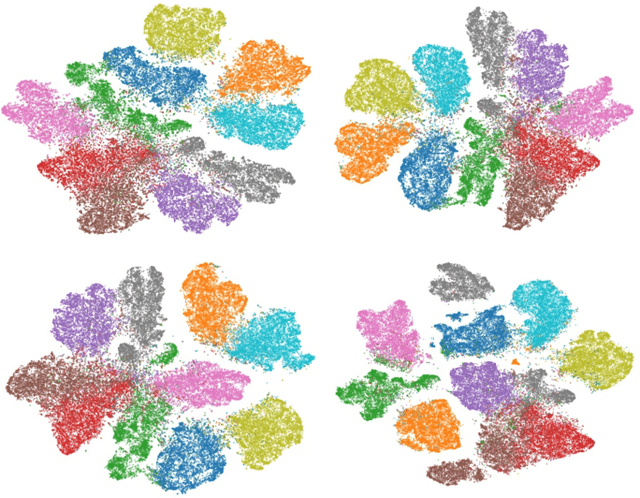
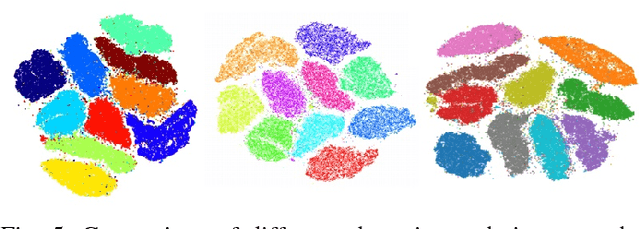
Abstract:Modern datasets and models are notoriously difficult to explore and analyze due to their inherent high dimensionality and massive numbers of samples. Existing visualization methods which employ dimensionality reduction to two or three dimensions are often inefficient and/or ineffective for these datasets. This paper introduces t-SNE-CUDA, a GPU-accelerated implementation of t-distributed Symmetric Neighbor Embedding (t-SNE) for visualizing datasets and models. t-SNE-CUDA significantly outperforms current implementations with 50-700x speedups on the CIFAR-10 and MNIST datasets. These speedups enable, for the first time, visualization of the neural network activations on the entire ImageNet dataset - a feat that was previously computationally intractable. We also demonstrate visualization performance in the NLP domain by visualizing the GloVe embedding vectors. From these visualizations, we can draw interesting conclusions about using the L2 metric in these embedding spaces. t-SNE-CUDA is publicly available at https://github.com/CannyLab/tsne-cuda
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge