Renske Gahrmann
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Evaluating glioma growth predictions as a forward ranking problem
Mar 22, 2021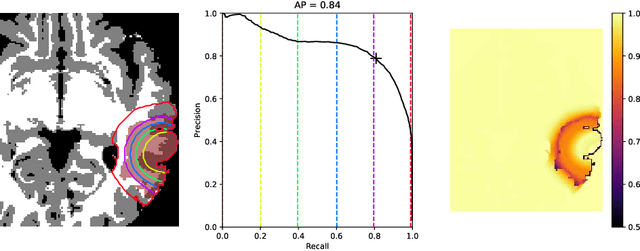
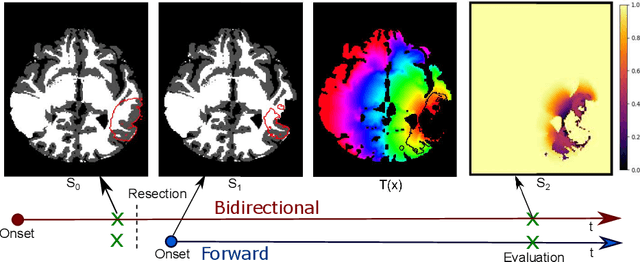
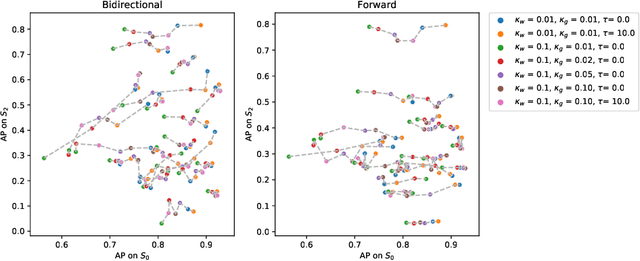
Abstract:The problem of tumor growth prediction is challenging, but promising results have been achieved with both model-driven and statistical methods. In this work, we present a framework for the evaluation of growth predictions that focuses on the spatial infiltration patterns, and specifically evaluating a prediction of future growth. We propose to frame the problem as a ranking problem rather than a segmentation problem. Using the average precision as a metric, we can evaluate the results with segmentations while using the full spatiotemporal prediction. Furthermore, by separating the model goodness-of-fit from future predictive performance, we show that in some cases, a better fit of model parameters does not guarantee a better the predictive power.
WHO 2016 subtyping and automated segmentation of glioma using multi-task deep learning
Oct 09, 2020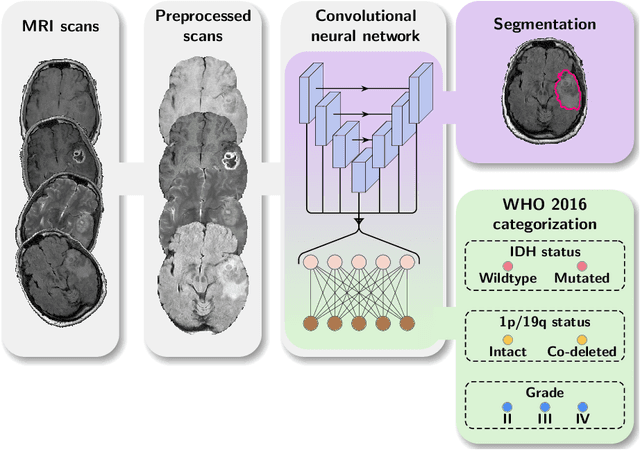
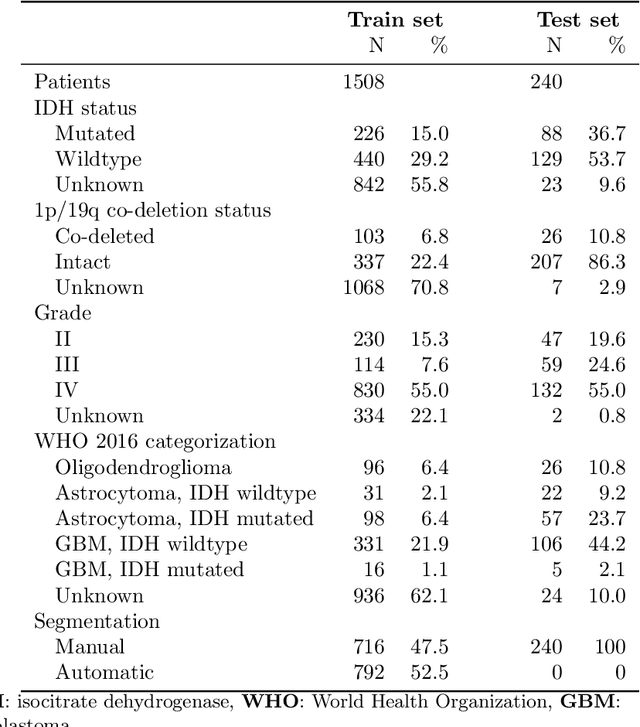
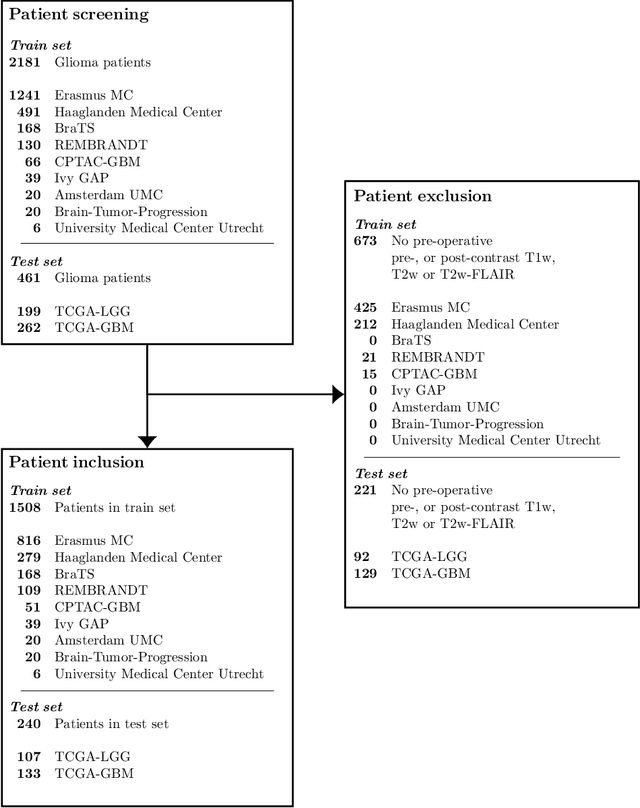
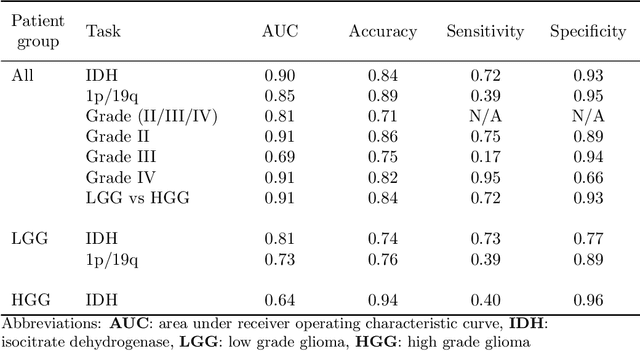
Abstract:Accurate characterization of glioma is crucial for clinical decision making. A delineation of the tumor is also desirable in the initial decision stages but is a time-consuming task. Leveraging the latest GPU capabilities, we developed a single multi-task convolutional neural network that uses the full 3D, structural, pre-operative MRI scans to can predict the IDH mutation status, the 1p/19q co-deletion status, and the grade of a tumor, while simultaneously segmenting the tumor. We trained our method using the largest, most diverse patient cohort to date containing 1508 glioma patients from 16 institutes. We tested our method on an independent dataset of 240 patients from 13 different institutes, and achieved an IDH-AUC of 0.90, 1p/19q-AUC of 0.85, grade-AUC of 0.81, and a mean whole tumor DICE score of 0.84. Thus, our method non-invasively predicts multiple, clinically relevant parameters and generalizes well to the broader clinical population.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge