Renat Sergazinov
A spectral method for multi-view subspace learning using the product of projections
Oct 24, 2024



Abstract:Multi-view data provides complementary information on the same set of observations, with multi-omics and multimodal sensor data being common examples. Analyzing such data typically requires distinguishing between shared (joint) and unique (individual) signal subspaces from noisy, high-dimensional measurements. Despite many proposed methods, the conditions for reliably identifying joint and individual subspaces remain unclear. We rigorously quantify these conditions, which depend on the ratio of the signal rank to the ambient dimension, principal angles between true subspaces, and noise levels. Our approach characterizes how spectrum perturbations of the product of projection matrices, derived from each view's estimated subspaces, affect subspace separation. Using these insights, we provide an easy-to-use and scalable estimation algorithm. In particular, we employ rotational bootstrap and random matrix theory to partition the observed spectrum into joint, individual, and noise subspaces. Diagnostic plots visualize this partitioning, providing practical and interpretable insights into the estimation performance. In simulations, our method estimates joint and individual subspaces more accurately than existing approaches. Applications to multi-omics data from colorectal cancer patients and nutrigenomic study of mice demonstrate improved performance in downstream predictive tasks.
GlucoBench: Curated List of Continuous Glucose Monitoring Datasets with Prediction Benchmarks
Oct 08, 2024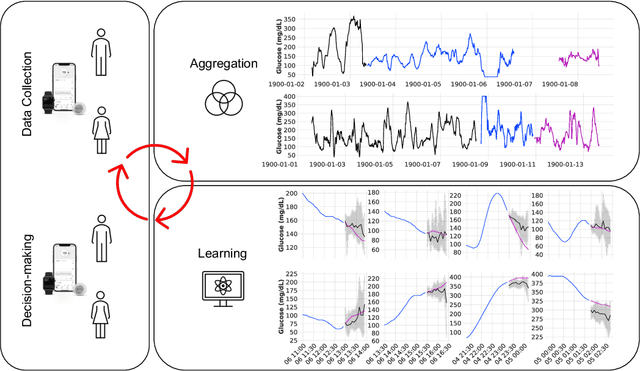

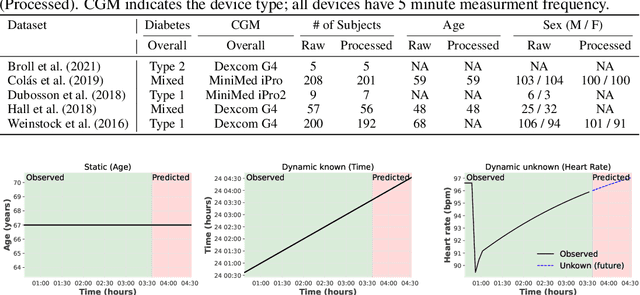
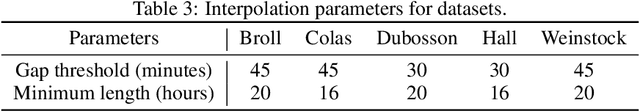
Abstract:The rising rates of diabetes necessitate innovative methods for its management. Continuous glucose monitors (CGM) are small medical devices that measure blood glucose levels at regular intervals providing insights into daily patterns of glucose variation. Forecasting of glucose trajectories based on CGM data holds the potential to substantially improve diabetes management, by both refining artificial pancreas systems and enabling individuals to make adjustments based on predictions to maintain optimal glycemic range.Despite numerous methods proposed for CGM-based glucose trajectory prediction, these methods are typically evaluated on small, private datasets, impeding reproducibility, further research, and practical adoption. The absence of standardized prediction tasks and systematic comparisons between methods has led to uncoordinated research efforts, obstructing the identification of optimal tools for tackling specific challenges. As a result, only a limited number of prediction methods have been implemented in clinical practice. To address these challenges, we present a comprehensive resource that provides (1) a consolidated repository of curated publicly available CGM datasets to foster reproducibility and accessibility; (2) a standardized task list to unify research objectives and facilitate coordinated efforts; (3) a set of benchmark models with established baseline performance, enabling the research community to objectively gauge new methods' efficacy; and (4) a detailed analysis of performance-influencing factors for model development. We anticipate these resources to propel collaborative research endeavors in the critical domain of CGM-based glucose predictions. {Our code is available online at github.com/IrinaStatsLab/GlucoBench.
SwitchTab: Switched Autoencoders Are Effective Tabular Learners
Jan 04, 2024Abstract:Self-supervised representation learning methods have achieved significant success in computer vision and natural language processing, where data samples exhibit explicit spatial or semantic dependencies. However, applying these methods to tabular data is challenging due to the less pronounced dependencies among data samples. In this paper, we address this limitation by introducing SwitchTab, a novel self-supervised method specifically designed to capture latent dependencies in tabular data. SwitchTab leverages an asymmetric encoder-decoder framework to decouple mutual and salient features among data pairs, resulting in more representative embeddings. These embeddings, in turn, contribute to better decision boundaries and lead to improved results in downstream tasks. To validate the effectiveness of SwitchTab, we conduct extensive experiments across various domains involving tabular data. The results showcase superior performance in end-to-end prediction tasks with fine-tuning. Moreover, we demonstrate that pre-trained salient embeddings can be utilized as plug-and-play features to enhance the performance of various traditional classification methods (e.g., Logistic Regression, XGBoost, etc.). Lastly, we highlight the capability of SwitchTab to create explainable representations through visualization of decoupled mutual and salient features in the latent space.
Gluformer: Transformer-Based Personalized Glucose Forecasting with Uncertainty Quantification
Sep 09, 2022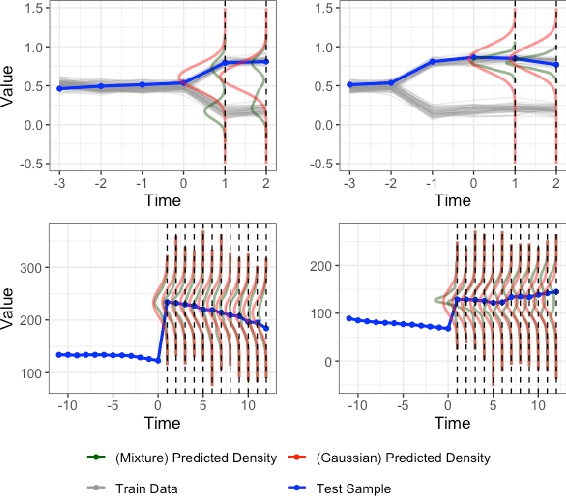
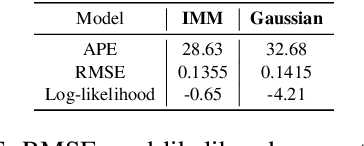
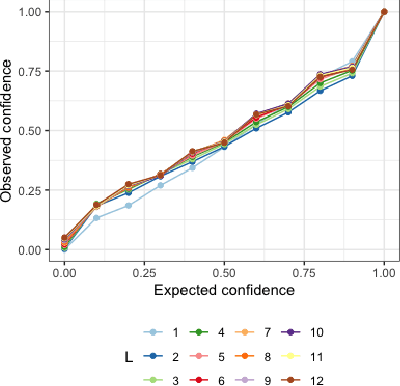
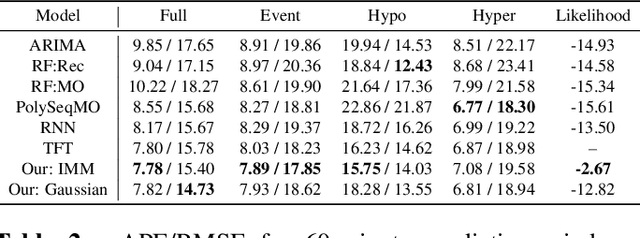
Abstract:Deep learning models achieve state-of-the art results in predicting blood glucose trajectories, with a wide range of architectures being proposed. However, the adaptation of such models in clinical practice is slow, largely due to the lack of uncertainty quantification of provided predictions. In this work, we propose to model the future glucose trajectory conditioned on the past as an infinite mixture of basis distributions (i.e., Gaussian, Laplace, etc.). This change allows us to learn the uncertainty and predict more accurately in the cases when the trajectory has a heterogeneous or multi-modal distribution. To estimate the parameters of the predictive distribution, we utilize the Transformer architecture. We empirically demonstrate the superiority of our method over existing state-of-the-art techniques both in terms of accuracy and uncertainty on the synthetic and benchmark glucose data sets.
Machine learning approach to force reconstruction in photoelastic materials
Oct 17, 2020



Abstract:Photoelastic techniques have a long tradition in both qualitative and quantitative analysis of the stresses in granular materials. Over the last two decades, computational methods for reconstructing forces between particles from their photoelastic response have been developed by many different experimental teams. Unfortunately, all of these methods are computationally expensive. This limits their use for processing extensive data sets that capture the time evolution of granular ensembles consisting of a large number of particles. In this paper, we present a novel approach to this problem which leverages the power of convolutional neural networks to recognize complex spatial patterns. The main drawback of using neural networks is that training them usually requires a large labeled data set which is hard to obtain experimentally. We show that this problem can be successfully circumvented by pretraining the networks on a large synthetic data set and then fine-tuning them on much smaller experimental data sets. Due to our current lack of experimental data, we demonstrate the potential of our method by changing the size of the considered particles which alters the exhibited photoelastic patterns more than typical experimental errors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge