Qing Pan
Community Detection with Heterogeneous Block Covariance Model
Dec 04, 2024Abstract:Community detection is the task of clustering objects based on their pairwise relationships. Most of the model-based community detection methods, such as the stochastic block model and its variants, are designed for networks with binary (yes/no) edges. In many practical scenarios, edges often possess continuous weights, spanning positive and negative values, which reflect varying levels of connectivity. To address this challenge, we introduce the heterogeneous block covariance model (HBCM) that defines a community structure within the covariance matrix, where edges have signed and continuous weights. Furthermore, it takes into account the heterogeneity of objects when forming connections with other objects within a community. A novel variational expectation-maximization algorithm is proposed to estimate the group membership. The HBCM provides provable consistent estimates of memberships, and its promising performance is observed in numerical simulations with different setups. The model is applied to a single-cell RNA-seq dataset of a mouse embryo and a stock price dataset. Supplementary materials for this article are available online.
CycleGAN with Dual Adversarial Loss for Bone-Conducted Speech Enhancement
Nov 02, 2021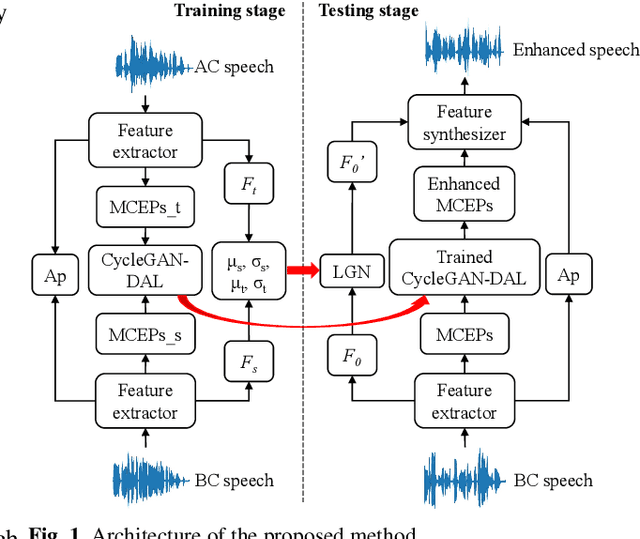
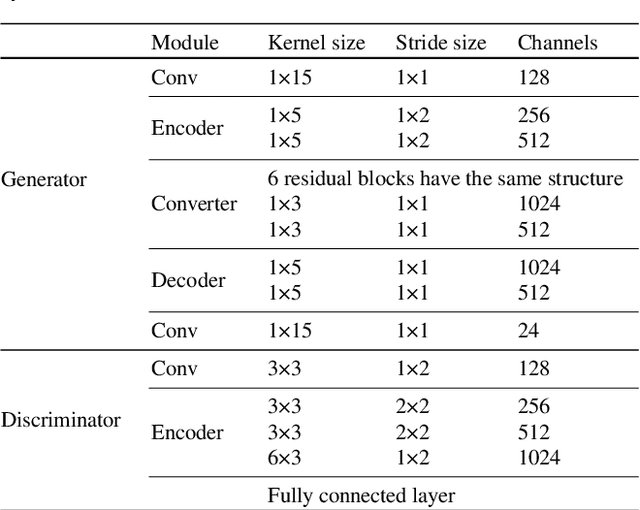
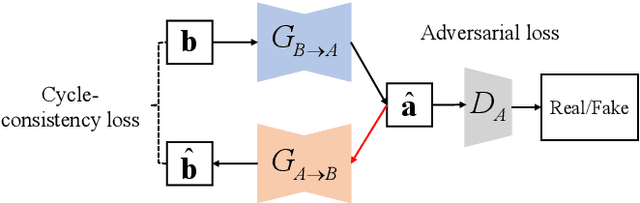
Abstract:Compared with air-conducted speech, bone-conducted speech has the unique advantage of shielding background noise. Enhancement of bone-conducted speech helps to improve its quality and intelligibility. In this paper, a novel CycleGAN with dual adversarial loss (CycleGAN-DAL) is proposed for bone-conducted speech enhancement. The proposed method uses an adversarial loss and a cycle-consistent loss simultaneously to learn forward and cyclic mapping, in which the adversarial loss is replaced with the classification adversarial loss and the defect adversarial loss to consolidate the forward mapping. Compared with conventional baseline methods, it can learn feature mapping between bone-conducted speech and target speech without additional air-conducted speech assistance. Moreover, the proposed method also avoids the oversmooth problem which is occurred commonly in conventional statistical based models. Experimental results show that the proposed method outperforms baseline methods such as CycleGAN, GMM, and BLSTM. Keywords: Bone-conducted speech enhancement, dual adversarial loss, Parallel CycleGAN, high frequency speech reconstruction
Logistic Regression Augmented Community Detection for Network Data with Application in Identifying Autism-Related Gene Pathways
Sep 07, 2018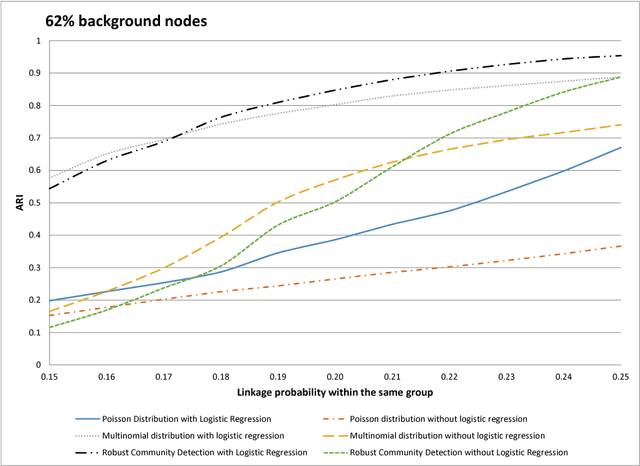
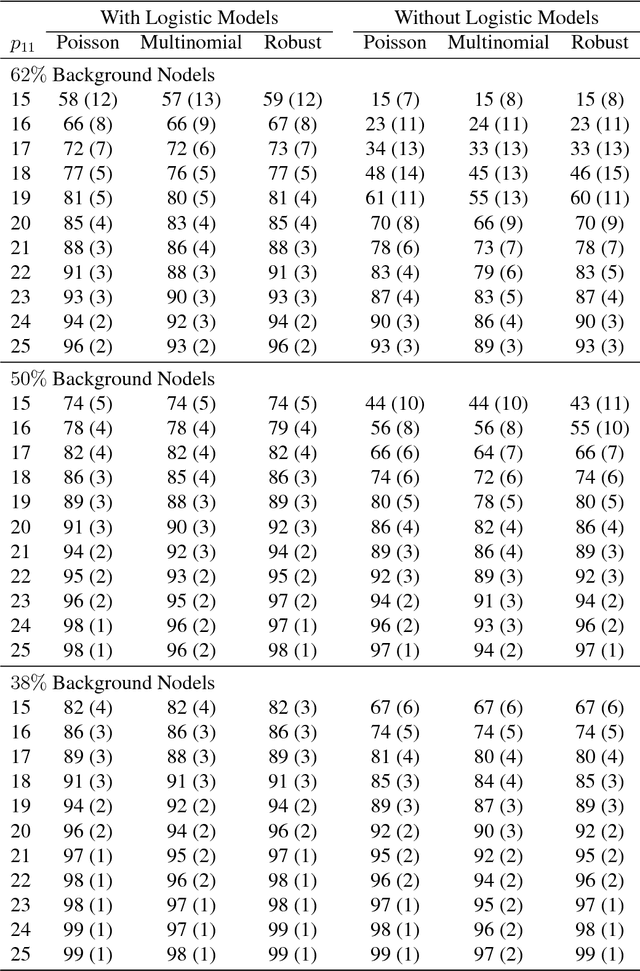

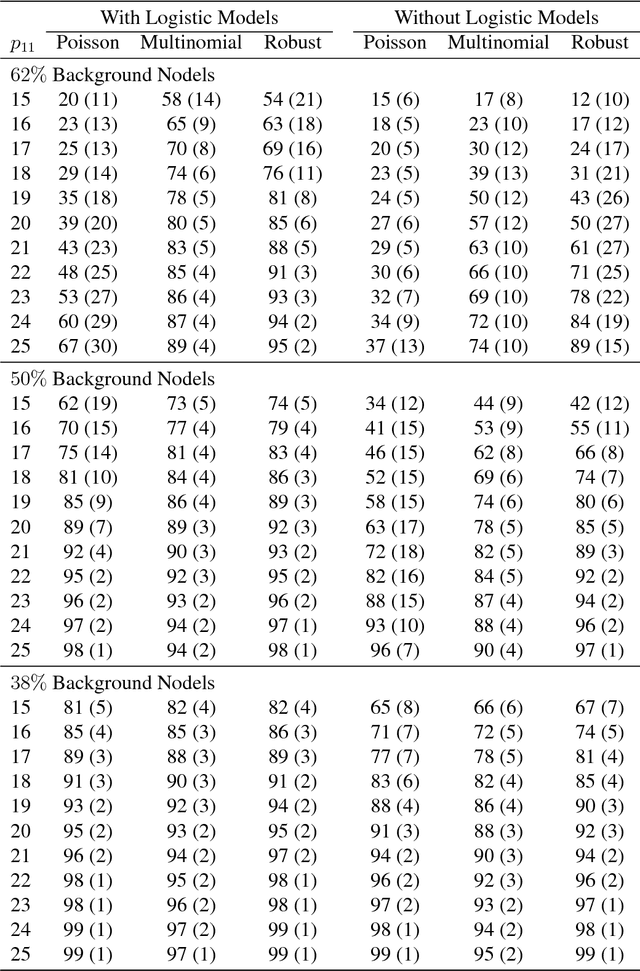
Abstract:When searching for gene pathways leading to specific disease outcomes, additional information on gene characteristics is often available that may facilitate to differentiate genes related to the disease from irrelevant background when connections involving both types of genes are observed and their relationships to the disease are unknown. We propose method to single out irrelevant background genes with the help of auxiliary information through a logistic regression, and cluster relevant genes into cohesive groups using the adjacency matrix. Expectation-maximization algorithm is modified to maximize a joint pseudo-likelihood assuming latent indicators for relevance to the disease and latent group memberships as well as Poisson or multinomial distributed link numbers within and between groups. A robust version allowing arbitrary linkage patterns within the background is further derived. Asymptotic consistency of label assignments under the stochastic blockmodel is proven. Superior performance and robustness in finite samples are observed in simulation studies. The proposed robust method identifies previously missed gene sets underlying autism related neurological diseases using diverse data sources including de novo mutations, gene expressions and protein-protein interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge