Qijian Chen
Multilevel Perception Boundary-guided Network for Breast Lesion Segmentation in Ultrasound Images
Oct 23, 2023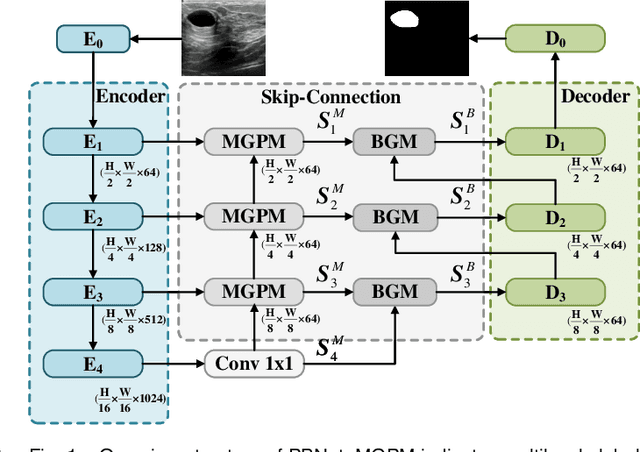
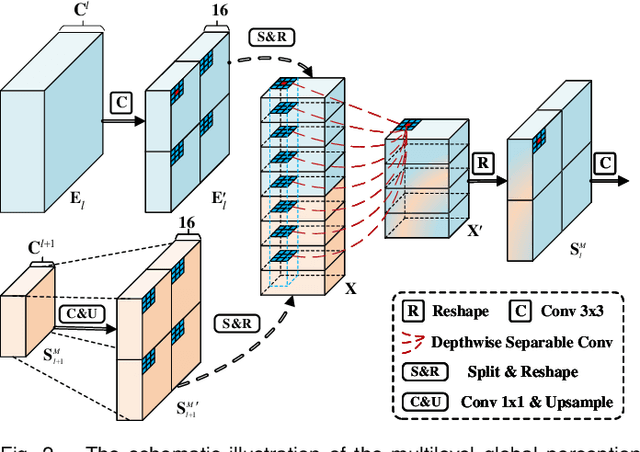
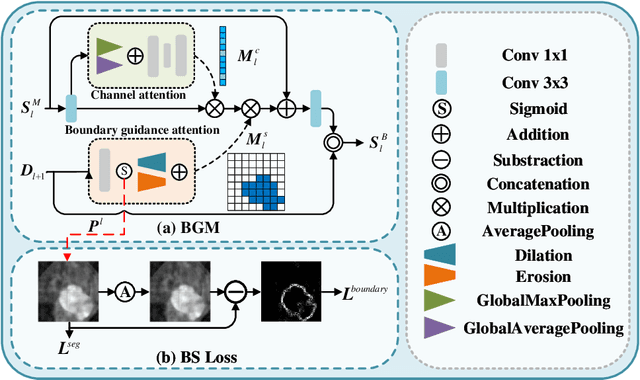
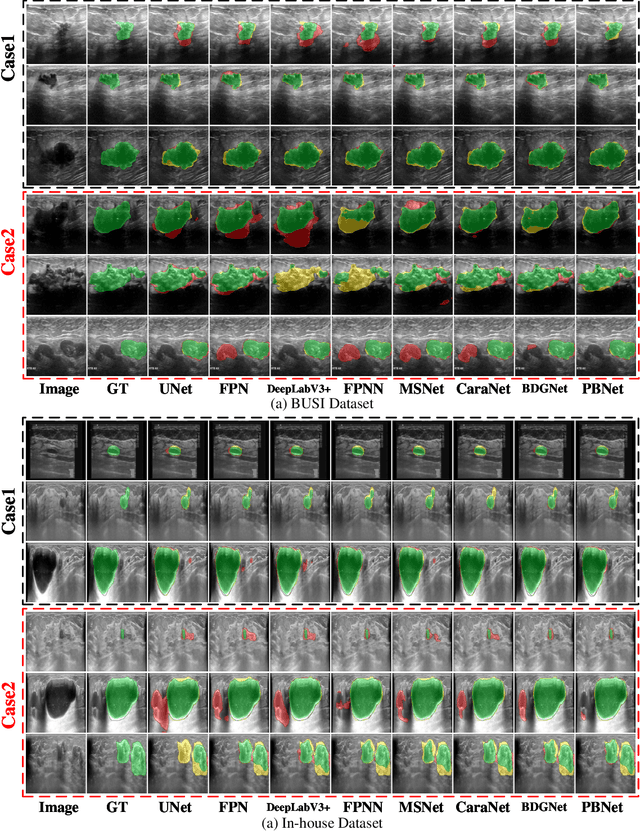
Abstract:Automatic segmentation of breast tumors from the ultrasound images is essential for the subsequent clinical diagnosis and treatment plan. Although the existing deep learning-based methods have achieved significant progress in automatic segmentation of breast tumor, their performance on tumors with similar intensity to the normal tissues is still not pleasant, especially for the tumor boundaries. To address this issue, we propose a PBNet composed by a multilevel global perception module (MGPM) and a boundary guided module (BGM) to segment breast tumors from ultrasound images. Specifically, in MGPM, the long-range spatial dependence between the voxels in a single level feature maps are modeled, and then the multilevel semantic information is fused to promote the recognition ability of the model for non-enhanced tumors. In BGM, the tumor boundaries are extracted from the high-level semantic maps using the dilation and erosion effects of max pooling, such boundaries are then used to guide the fusion of low and high-level features. Moreover, to improve the segmentation performance for tumor boundaries, a multi-level boundary-enhanced segmentation (BS) loss is proposed. The extensive comparison experiments on both publicly available dataset and in-house dataset demonstrate that the proposed PBNet outperforms the state-of-the-art methods in terms of both qualitative visualization results and quantitative evaluation metrics, with the Dice score, Jaccard coefficient, Specificity and HD95 improved by 0.70%, 1.1%, 0.1% and 2.5% respectively. In addition, the ablation experiments validate that the proposed MGPM is indeed beneficial for distinguishing the non-enhanced tumors and the BGM as well as the BS loss are also helpful for refining the segmentation contours of the tumor.
CNN-Based Invertible Wavelet Scattering for the Investigation of Diffusion Properties of the In Vivo Human Heart in Diffusion Tensor Imaging
Dec 17, 2019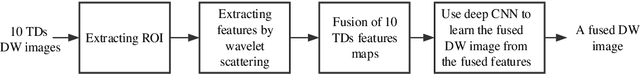
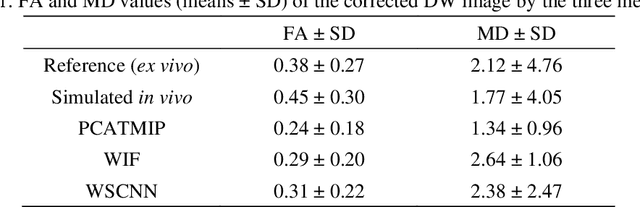
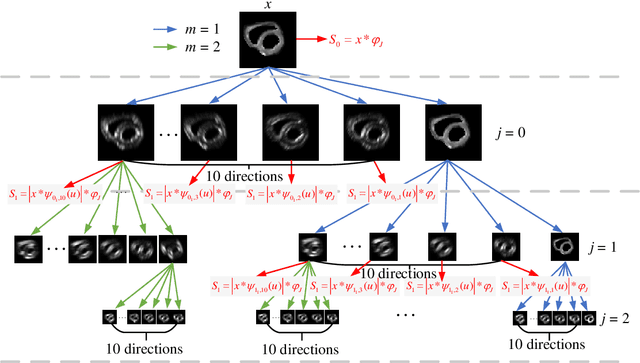
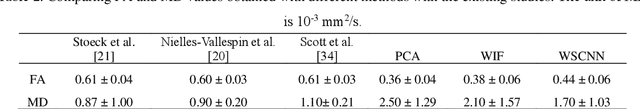
Abstract:In vivo diffusion tensor imaging (DTI) is a promising technique to investigate noninvasively the fiber structures of the in vivo human heart. However, signal loss due to motions remains a persistent problem in in vivo cardiac DTI. We propose a novel motion-compensation method for investigating in vivo myocardium structures in DTI with free-breathing acquisitions. The method is based on an invertible Wavelet Scattering achieved by means of Convolutional Neural Network (WSCNN). It consists of first extracting translation-invariant wavelet scattering features from DW images acquired at different trigger delays and then mapping the fused scattering features into motion-compensated spatial DW images by performing an inverse wavelet scattering transform achieved using CNN. The results on both simulated and acquired in vivo cardiac DW images showed that the proposed WSCNN method effectively compensates for motion-induced signal loss and produces in vivo cardiac DW images with better quality and more coherent fiber structures with respect to existing methods, which makes it an interesting method for measuring correctly the diffusion properties of the in vivo human heart in DTI under free breathing.
Glioma Grade Predictions using Scattering Wavelet Transform-Based Radiomics
May 23, 2019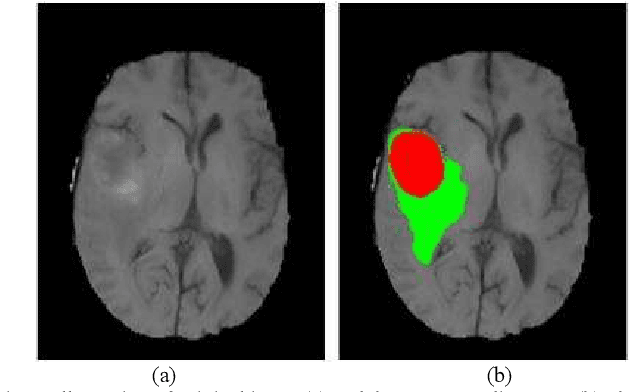
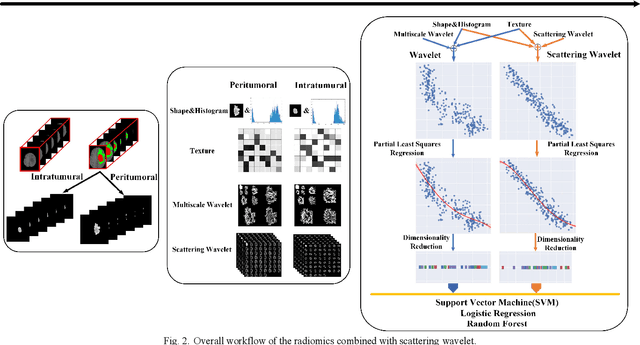
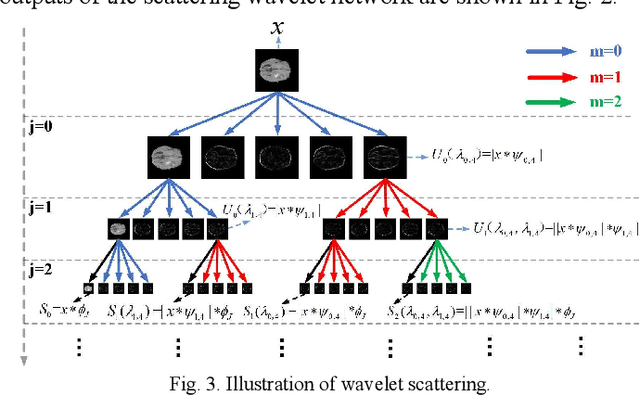
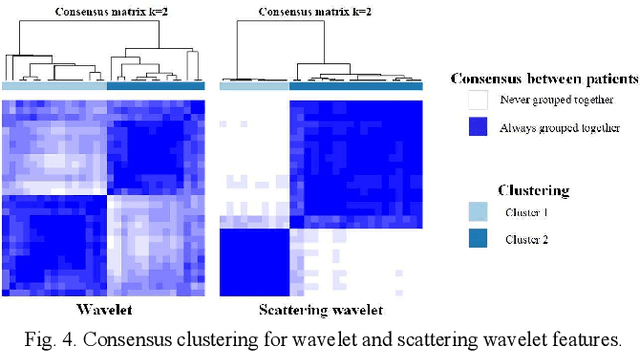
Abstract:Glioma grading before the surgery is very critical for the prognosis prediction and treatment plan making. In this paper, we present a novel scattering wavelet-based radiomics method to predict noninvasively and accurately the glioma grades. The multimodal magnetic resonance images of 285 patients were used, with the intratumoral and peritumoral regions well labeled. The wavelet scattering-based features and traditional radiomics features were firstly extracted from both intratumoral and peritumoral regions respectively. The support vector machine (SVM), logistic regression (LR) and random forest (RF) were then trained with 5-fold cross validation to predict the glioma grades. The prediction obtained with different features was finally evaluated in terms of quantitative metrics. The area under the receiver operating characteristic curve (AUC) of glioma grade prediction based on scattering wavelet features was up to 0.99 when considering both intratumoral and peritumoral features in multimodal images, which increases by about 17% compared to traditional radiomics. Such results shown that the local invariant features extracted from the scattering wavelet transform allows improving the prediction accuracy for glioma grading. In addition, the features extracted from peritumoral regions further increases the accuracy of glioma grading.
Convolutional Restricted Boltzmann Machine Based-Radiomics for Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy in Breast Cancer
May 23, 2019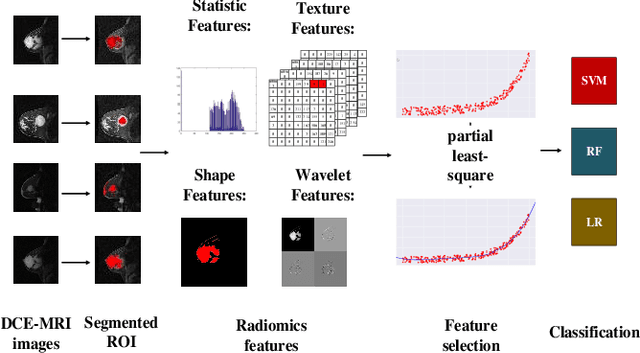


Abstract:We proposed a novel convolutional restricted Boltzmann machine CRBM-based radiomic method for predicting pathologic complete response (pCR) to neoadjuvant chemotherapy treatment (NACT) in breast cancer. The method consists of extracting semantic features from CRBM network, and pCR prediction. It was evaluated on the dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) data of 57 patients and using the area under the receiver operating characteristic curve (AUC). Traditional radiomics features and the semantic features learned from CRBM network were extracted from the images acquired before and after the administration of NACT. After the feature selection, the support vector machine (SVM), logistic regression (LR) and random forest (RF) were trained to predict the pCR status. Compared to traditional radiomic methods, the proposed CRBM-based radiomic method yielded an AUC of 0.92 for the prediction with the images acquired before and after NACT, and an AUC of 0.87 for the pretreatment prediction, which was increased by about 38%. The results showed that the CRBM-based radiomic method provided a potential means for accurately predicting the pCR to NACT in breast cancer before the treatment, which is very useful for making more appropriate and personalized treatment regimens.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge